UGENE
 | |
 | |
| Developer(s) | Unipro |
|---|---|
| Stable release | 1.16.1 / 18 March 2015 |
| Written in | C++, Qt |
| Operating system | Cross-platform |
| Available in | English, Russian, Czech, Chinese |
| Type | Bioinformatics toolkit |
| License | GPL |
| Website |
ugene |
UGENE is free open-source bioinformatics software.[1][2] It works on a desktop computer with Windows, Mac OS X or Linux.
UGENE helps biologists to analyze various biological data, such as sequences, annotations, multiple alignments, phylogenetic trees, NGS assemblies, and others. The data can be stored both locally (on a personal computer) and on a shared storage (e.g. a lab database).
UGENE integrates dozens of well-known biological tools and algorithms, as well as original tools in context of genomics, evolutionary biology, virology and other branches of life science. UGENE provides a graphical interface for the pre-built tools so biologists without programming skills can access those tools more easily.
Using UGENE Workflow Designer, it is possible to streamline a multi-step analysis. The workflow consists of blocks such as data readers, blocks executing embedded tools/algorithms and data writers. Blocks can be created with command line tools or a script. A set of sample workflows is available in the Workflow Designer (for annotating sequences, conversion of data formats, NGS data analysis, etc.)
Besides the graphical interface, UGENE also provides a command-line interface. A worklow made by the Workflow Designer can be executed using the command-line interface.
In order to provide better performance, UGENE utilizes multicore CPUs and GPUs to optimize a few algorithms.[3][4]
Key features
The software supports the following features:
- Creating, editing and annotating nucleic acid and protein sequences
- Fast search in a sequence
- Multiple sequence alignment: ClustalW, ClustalO, MUSCLE, Kalign, MAFFT, T-Coffee
- Creating and using a shared storage (e.g.lab database)
- Search through online databases: NCBI, PDB, UniProtKB/Swiss-Prot, UniProtKB/TrEMBL, DAS servers
- Local and NCBI Genbank BLAST search
- Open reading frame finder
- Restriction enzyme finder with integrated REBASE restriction enzymes list
- Integrated Primer3 package for PCR primer design
- Plasmid construction and annotation
- Cloning in silico by designing of cloning vectors
- Genome mapping of short reads with Bowtie, BWA and UGENE Genome Aligner
- Visualization of next generation sequencing data (BAM files) using UGENE Assembly Browser
- Variant calling with SAMtools
- RNA-seq data analysis with Tuxedo pipeline (TopHat, Cufflinks, etc.)
- ChIP-seq data analysis with Cistrome pipeline (MACS, CEAS, etc.)
- Raw NGS data processing
- HMMER2 and HMMER3 packages integration
- Chromatogram viewer
- Search for transcription factor binding sites (TFBS) with weight matrix and SITECON algorithms
- Search for direct, inverted and tandem repeats in DNA sequences
- Local sequence alignment with optimized Smith-Waterman algorithm
- Building (using integrated PHYLIP Neighbor Joining, MrBayes or PhyML Maximum Likelihood) and editing phylogenetic trees
- Combining various algorithms into custom workflows with UGENE Workflow Designer
- Contigs assembly with CAP3
- 3D structure viewer for files in PDB and MMDB formats, anaglyph view support
- Protein secondary structure prediction with GOR IV and PSIPRED algorithms
- Constructing dotplots for nucleic acid sequences
- mRNA alignment with Spidey
- Search for complex signals with ExpertDiscovery[5]
- Search for a pattern of various algorithms' results in a nucleic acid sequence with UGENE Query Designer
- PCR in silico for primer designing and mapping
- Spade de novo assembler
Sequence View
The Sequence View is used to visualize, analyze and modify nucleic acid or protein sequences. Depending on the sequence type and the options selected, the following views can be present in the Sequence View window:
- 3D structure view
- Circular view
- Chromatogram view
- Graphs View (GC-content, AG-content and other)
- Dotplot view
Alignment Editor
The Alignment Editor allows working with multiple nucleic acid or protein sequences - aligning them, editing the alignment, analyzing it, storing the consensus sequence, building a phylogenetic tree and so on.
Phylogenetic Tree Viewer
The Phylogenetic Tree Viewer helps to visualize and edit phylogenetic trees. It is possible to synchronize a tree with the corresponding multiple alignment used to build the tree.
Assembly Browser
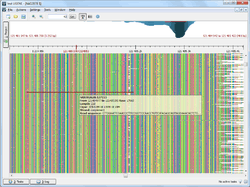
Assembly Browser project was started in 2010 as an entry for Illumina iDEA Challenge 2011. The Assembly Browser allows a user to visualize and browse large (up to hundreds of millions of short reads) next generation sequence assemblies. It supports SAM, BAM (which is the binary version of SAM) and ACE formats. Before browsing assembly data in UGENE, an input file is converted to a UGENE database file automatically. This approach has its pros and cons. The cons are that the conversion may take time for a large file and there should be enough disk space to store the database. On the other hand, this allows to view the whole assembly, navigate in it and go to well-covered regions rather rapidly.
Workflow Designer
UGENE Workflow Designer allows creating and running complex computational workflow schemas.[6]
The distinguishing feature of the UGENE Workflow Designer in comparison with other bioinformatics workflow management systems is that workflows in UGENE are executed on a local computer. It helps to avoid data transfer issues, whereas other tools’ reliance on remote file storage and internet connectivity does not.
The elements that a workflow consists of correspond to the bulk of algorithms integrated into UGENE. Using the Workflow Designer one can also create custom workflow elements. The elements can be based on a command-line tool or a script.
Workflows are stored in a special text format. This allows transferring the workflows to other users and reusing them.
A workflow can be run using the graphical interface or launched from the command line. The graphical interface additionally allows controlling the workflow execution, storing the parameters, and so on.
There is an embedded library of workflow samples for converting, filtering and annotating of data. Particularly, there are several pipelines for analysis of NGS data developed in collaboration with NIH NIAID. A wizard is available for each workflow sample.
Supported biological data formats
- Sequences and annotations: FASTA (.fa), GenBank (.gb), EMBL (.emb), GFF (.gff)
- Multiple sequence alignments: Clustal (.aln), MSF (.msf), Stockholm (.sto), Nexus (.nex)
- 3D structures: PDB (.pdb), MMDB (.prt)
- Chromatograms: ABIF (.abi), SCF (.scf)
- Short reads: Sequence Alignment/Map(SAM) (.sam), binary version of SAM (.bam), ACE (.ace), FASTQ (.fastq)
- Phylogenetic trees: Newick (.nwk), PHYLIP (.phy)
- Other formats: Bairoch (enzymes info), HMM (HMMER profiles), PWM and PFM (position matrices), SNP and VCF4 (genome variations)
Release cycle
UGENE is primarily developed by Unipro LLC with headquarters in Akademgorodok of Novosibirsk, Russia. Each iteration lasts about 1–2 months, followed by a new release comes out. One can also download a development snapshot of the software.
The features to be included in each release are mostly initiated by users.
See also
- Sequence alignment software
- Bioinformatics
- Computational biology
- List of open source bioinformatics software
Related software
- Gene Designer
- Galaxy
- Vector NTI
- Geneious
- MacVector
References
- ↑ Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team (2012). "Unipro UGENE: a unified bioinformatics toolkit". Bioinformatics 28 (8): 1166–7. doi:10.1093/bioinformatics/bts091. PMID 22368248.
- ↑ Fursov, M.; Novikova, O. (2008). "Multitasking software system for DNA analysis". Proceedings of the Sixth International Conference on Bioinformatics of Genome Regulation and Structure 1: 78. ISBN 978-5-91291-005-0.
- ↑ Fursov, M. Y.; Oshchepkov, D. Y; Novikova, O. S. (2009). "UGENE: interactive computational schemes for genome analysis". Proceedings of the Fifth Moscow International Congress on Biotechnology 3: 14–15. ISBN 5-7237-0372-2.
- ↑ Efremov, I. E.; Fursov, M. Y; Danilova, Yu. E. (2009). "UGENE: high performance genome analysis suite". Proceedings of the Fifth Moscow International Congress on Biotechnology 2: 405–406. ISBN 5-7237-0372-2.
- ↑ Vaskin, Y.; Khomicheva, I.; Ignatieva, E.; Vityaev, E.; (2012). "ExpertDiscovery and UGENE integrated system for intelligent analysis of regulatory regions of genes". In Silico Biology 11 (3-4): 97–108. doi:10.3233/ISB-2012-0448. PMID 22935964.
- ↑ Fursov, M. Y.; Varlamov, A. (2009). "UGENE - A practical approach for complex computational analysis in molecular biology". Proceedings of the 10th Annual Bioinformatics Open Source Conference: 7.