Translocon
The translocon (commonly known as a translocator or translocation channel) is a complex of proteins associated with the translocation of polypeptides across membranes.[1] In eukaryotes the term translocon most commonly refers to the complex that transports nascent polypeptides with a targeting signal sequence into the interior (cisternal or lumenal) space of the endoplasmic reticulum (ER) from the cytosol. This translocation process requires the protein to cross a hydrophobic lipid bilayer. The same complex is also used to integrate nascent proteins into the membrane itself (membrane proteins). In prokaryotes, a similar protein complex transports polypeptides across the plasma membrane or integrates membrane proteins.[2] Bacterial pathogens can also assemble other translocons in their host membranes, allowing them to export virulence factors into their target cells.[3]
The prokaryotic translocon
The bacterial translocation channel is formed by a trimeric protein complex called SecYEG. It consists of the subunits SecY, SecE, and SecG. The structure of the archaeal homolog of this complex has been determined by X-ray crystallography.[4] There has been a long-standing debate about the oligomeric state of the complex in the cell. Most recently, the monomeric form seems to be favored by most researchers.[5]
The ER-translocon
The translocon protein complexes are formed from Sec proteins.[6]
The translocon complex consists of several large protein complexes. The central element is the translocation channel itself, the heterotrimer Sec61. Further known components of the translocon complex are the oligosaccharyl transferase complex, the TRAP complex, and the membrane protein TRAM. For further components, such as signal peptidase complex and the SRP receptor it is not clear to what extent they only associate transiently to the translocon complex.
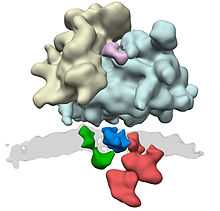
Proteins due to be translocated to the endoplasmic reticulum are recognized by the signal-recognition particle (SRP), which halts translation of the polypeptide by the ribosome while it attaches the ribosome to the SRP receptor on the endoplasmic reticulum. This recognition event is based upon a specific N-terminal signal sequence that is in the first few codons of the polypeptide to be synthesised.
The current model of protein translocation is that the translocon (translocator) acts as a channel through the hydrophobic membrane of the endoplasmic reticulum (after the SRP has dissociated and translation is continued). The emerging polypeptide is threaded through the channel as an unfolded string of amino acids. Once translation is finished a signal peptidase cleaves off the short signal peptide from the nascent protein leaving the polypeptide free in the interior of the endoplasmic reticulum.
The translocon can also translocate and integrate membrane proteins in the correct orientation into the membrane of the endoplasmic reticulum. The mechanism of this process is not fully understood, but involves the recognition and processing by the translocon of hydrophobic stretches in the amino acid sequence which are destined to become transmembrane helices.
The ER-retrotranslocon
Translocators can also move polypeptides (such as damaged proteins targeted for proteasomes) from the cisternal space of the endoplasmic reticulum to the cytosol. ER-proteins are degraded in the cytosol by the 26S proteasome, a process known as Endoplasmic-reticulum-associated protein degradation, and therefore have to be transported by an appropriate channel. This retrotranslocon is still enigmatic.
It was initially believed that the Sec61 channel is responsible for this retrograde transport, implying that transport through Sec61 is not always unidirectional but also can be bidirectional.[8] However, the structure of Sec61 does not support this view and several different proteins have been suggested to be responsible for transport from the ER lumen into the cytosol.[9]
See also
References
- ↑ Johnson, A.E.; van Waes, M.A. (1999). "The translocon: a dynamic gateway at the ER membrane". Annu. Rev. Cell Dev. Biol. doi:10.1146/annurev.cellbio.15.1.799.
- ↑ Gold VA, Duong F, Collinson I (2007). "Structure and function of the bacterial Sec translocon". Mol. Membr. Biol. 24 (5–6): 387–94. doi:10.1080/09687680701416570. PMID 17710643.
- ↑ Mueller CA, Broz P, Cornelis GR (June 2008). "The type III secretion system tip complex and translocon". Mol. Microbiol. 68 (5): 1085–95. doi:10.1111/j.1365-2958.2008.06237.x. PMID 18430138.
- ↑ "X-ray structure of a protein-conducting channel" 427 (6969). January 2004. pp. 36–44. doi:10.1038/nature02218. PMID 14661030.
- ↑ Kedrov, Alexej (2011). "A single copy of SecYEG is sufficient for preprotein translocation". The EMBO Journal 30 (21): 4387–4397. doi:10.1038/emboj.2011.314.
- ↑ Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. (1991). "Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex". Nature 349 (6312): 806–8. doi:10.1038/349806a0. PMID 2000150.
- ↑ Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Förster F (2014). "Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon". Nat. Commun. (5): 3072. doi:10.1038/ncomms4072.
- ↑ Römisch K (December 1999). "Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane". J. Cell. Sci. 112 (23): 4185–91. PMID 10564637.
- ↑ Hampton, R. Y.; Sommer, T. "Finding the will and the way of ERAD substrate retrotranslocation". Current opinion in cell biology. doi:10.1016/j.ceb.2012.05.010.