Synthetic biological circuit
Synthetic biological circuits are an application of synthetic biology where biological parts inside a cell are designed to perform logical functions mimicking those observed in electronic circuits. The applications range from simply inducing production to adding a measurable element, like GFP, to an existing natural biological circuit, to implementing completely new systems of many parts.[1]
The goal of synthetic biology is to generate an array of tunable and characterized parts, or modules, with which any desirable synthetic biological circuit can be easily designed and implemented.[2] These circuits can serve as a method to modify cellular functions, create cellular responses to environmental conditions, or influence cellular development. By implementing rational, controllable logic elements in cellular systems, researchers can use living systems as engineered "machines" to perform a vast range of useful functions.[1]
History
The first natural gene circuit studied in detail was the lac operon. In studies of diauxic growth of E. coli on two-sugar media, Jacques Monod and Francois Jacob discovered that E.coli preferentially consumes the more easily processed glucose before switching to lactose metabolism. They discovered that the mechanism that controlled the metabolic "switching" function was a two-part control mechanism on the lac operon. When lactose is present in the cell the enzyme β-galactosidase is produced to convert lactose into glucose or galactose. When lactose is absent in the cell the lac repressor inhibits the production of the enzyme β-galactosidase to prevent any inefficient processes within the cell.
The lac operon is used in the biotechnology industry for production of recombinant proteins for therapeutic use. The gene or genes for producing an exogenous protein are placed on a plasmid under the control of the lac promoter. Initially the cells are grown in a medium that does not contain lactose or other sugars, so the new genes are not expressed. Once the cells reach a certain point in their growth, Isopropyl_β-D-1-thiogalactopyranoside (IPTG) is added. IPTG, a molecule similar to lactose, but with a sulfur bond that is not hydrolyzable so that E. Coli does not digest it, is used to activate or "induce" the production of the new protein. Once the cells are induced, it is difficult to remove IPTG from the cells and therefore it is difficult to stop expression.
An early example of a synthetic biological circuit was published in Nature in 2000 by Tim Gardner, Charles Cantor, and Jim Collins working at Boston University. They were able to create a "bistable" switch in E. Coli. The switch is turned on by heating the culture of bacteria and turned off by addition of IPTG. They used GFP as a reporter for their system.[3]
Currently, synthetic circuits are a burgeoning area of research in systems biology with more publications detailing synthetic biological circuits published every year.[4] There has been significant interest in encouraging education and outreach as well: the International Genetically Engineered Machines Competition[5] manages the creation and standardization of BioBrick parts as a means to allow undergraduate and high school students to design their own synthetic biological circuits.
Interest and goals
Both immediate and long term applications exist for the use of synthetic biological circuits, including different applications for metabolic engineering, and synthetic biology. Those demonstrated successfully include pharmaceutical production,[6] and fuel production.[7] However methods involving direct genetic introduction are not inherently effective without invoking the basic principles of synthetic cellular circuits. For example, each of these successful systems employs a method to introduce all-or-none induction or expression. This is a biological circuit where a simple repressor or promoter is introduced to facilitate creation of the product, or inhibition of a competing pathway. However, with the limited understanding of cellular networks and natural circuitry, implementation of more robust schemes with more precise control and feedback is hindered. Therein lies the immediate interest in synthetic cellular circuits.
Development in understanding cellular circuitry can lead to exciting new modifications, such as cells which can respond to environmental stimuli. For example cells could be developed that signal toxic surroundings and react by activating pathways used to degrade the perceived toxin.[8] To develop such a cell, it is necessary to create a complex synthetic cellular circuit which can respond appropriately to a given stimuli.
Given synthetic cellular circuits represent a form of control for cellular activities, it can be reasoned that with complete understanding of cellular pathways, "plug and play"[1] cells with well defined genetic circuitry can be engineered. It is widely believed that if a proper toolbox of parts is generated,[9] synthetic cells can be developed implementing only the pathways necessary for cell survival reproduction. From this cell, to be thought of as a minimal genome cell, one can add pieces from the toolbox to create a well defined pathway with appropriate synthetic circuitry for an effective feedback system. Because of the basic ground up construction method, and the proposed database of mapped circuitry pieces, techniques mirroring those used to model computer or electronic circuits can be used to redesign cells and model cells for easy troubleshooting and predictive behavior and yields.
Example circuits
Logical operators
List of currently published circuits:
- "Repressilator"
- Toggle-switch
- Mammalian tunable synthetic oscillator
- Bacterial tunable synthetic oscillator
- Coupled bacterial oscillator
- Globally coupled bacterial oscillator


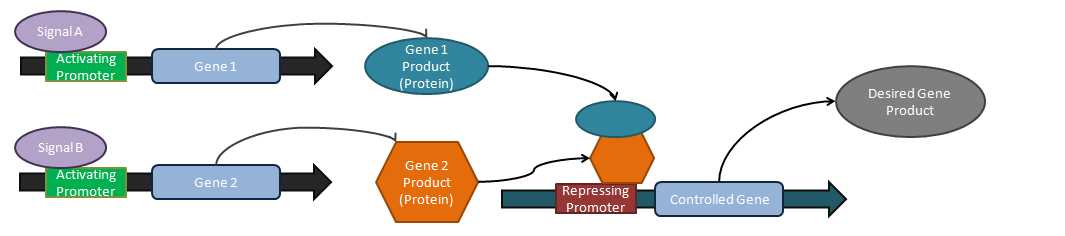
Engineered systems
Engineered systems are the result of implementation of combinations of different logical control mechanisms. Gardner et al. used a cascading response of multiple control units to create an implementation of a toggle switch capable of controlling metabolism by a stepwise function.[3] Elowitz et al. and Fung et al. created oscillatory circuits that use multiple self-regulating mechanisms to create a time-dependent oscillation of gene product expression.[12][13] A limited counting mechanism was implemented by a pulse-controlled gene cascade[14] and application of logic elements enables genetic "programming" of cells as in the research of Tabor et al., which synthesized a photosensitive bacterial edge detection program.[15]
References
- ↑ 1.0 1.1 1.2 H. Kobayashi, M. Kærn, M. Araki, K. Chung, T. S. Gardner, C. R. Cantor, J. J. Collins, Programmable cells: Interfacing natural and engineered gene networks. PNAS June 1, 2004 vol. 101 no. 22 8414-8419
- ↑ "Synthetic Biology: FAQ". SyntheticBiology.org. Retrieved 21 December 2011.
- ↑ 3.0 3.1 Gardner, T.s., Cantor, C.R., Collins, J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339-342 (20 January 2000).
- ↑ Priscilla E. M. Purnick & Ron Weis. The second wave of synthetic biology: from modules to systems. Nature Reviews Molecular Cell Biology 10, 410-422 (June 2009) | doi:10.1038/nrm2698
- ↑ International Genetically Engineered Machines (iGem) http://igem.org/Main_Page
- ↑ D.-K. Ro, E.M. Paradise, M. Ouellet, K.J. Fisher, K.L. Newman, J.M. Ndungu, K.A. Ho, R.A. Eachus, T.S. Ham, J. Kirby, M.C.Y. Chang, S.T. Withers, Y. Shiba, R. Sarpong and J.D. Keasling, Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature, 440 (2006), pp. 940–943
- ↑ J.L. Fortman, S. Chhabra, A. Mukhopadhyay, H. Chou, T.S. Lee, E. Steen and J.D. Keasling, Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol, 26 (2008), pp. 375–381.
- ↑ J.D. Keasling, Synthetic biology for synthetic chemistry. ACS Chem Biol, 3 (2008), pp. 64–76.
- ↑ Julius B Lucks, Lei Qi, Weston R Whitaker, Adam P Arkin, Toward scalable parts families for predictable design of biological circuits, Current Opinion in Microbiology, Volume 11, Issue 6, December 2008, Pages 567-573, ISSN 1369-5274, 10.1016/j.mib.2008.10.002. (http://www.sciencedirect.com/science/article/pii/S1369527408001410)
- ↑ 10.0 10.1 10.2 Silva-Rocha, R., de Lorenzo, V. Mining logic gates in prokaryotic transcriptional regulation networks. FEBS Letters, April 9, 2008 vol. 582 no. 8 1237-1244
- ↑ 11.0 11.1 11.2 Buchler, N.E., Gerland, U., Hwa, T. On schemes of combinatorial transcription logic. PNAS April 29, 2003 vol. 100 no. 9 5136-5141
- ↑ Elowitz, M.B., Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335-338 (20 January 2000).
- ↑ Fung, E., Wong, W.W., Suen, J.K., Bulter, T., Lee, S., Liao, J.C. A synthetic gene–metabolic oscillator. Nature 435, 118-122 (5 May 2005).
- ↑ Friedland, A.E., Lu, T.K, Wang, X., Shi, D., Church, G., Collins, J.J. Synthetic Gene Networks That Count. Science vol. 324 no. 5931, 1199-1202 (29 May 2009).
- ↑ Tabor, J.J., Salis, H.M., Simpson, Z.B., Chevalier, A.A., Levskaya, A., Marcotte, E.M., Voigt, C.A., Ellington, A.D. A Synthetic Edge Detection Program. Cell 137, 1272-1281 (June 26, 2009).