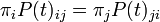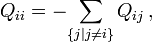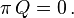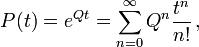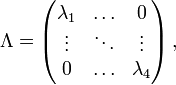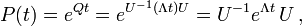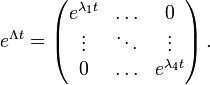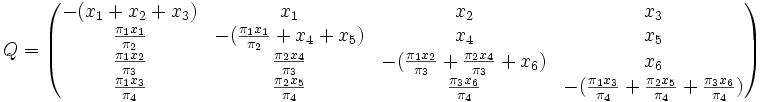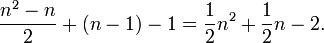Substitution model
In biology, a substitution model describes the process from which a sequence of characters changes into another set of traits. For example, in cladistics, each position in the sequence might correspond to a property of a species which can either be present or absent. The alphabet could then consist of "0" for absence and "1" for presence. Then the sequence 00110 could mean, for example, that a species does not have feathers or lay eggs, does have fur, is warm-blooded, and cannot breathe underwater. Another sequence 11010 would mean that a species has feathers, lays eggs, does not have fur, is warm-blooded, and cannot breathe underwater. In phylogenetics, sequences are often obtained by firstly obtaining a nucleotide or protein sequence alignment, and then taking the bases or amino acids at corresponding positions in the alignment as the characters. Sequences achieved by this might look like AGCGGAGCTTA and GCCGTAGACGC.
Substitution models are used for a number of things:
- Constructing evolutionary trees in phylogenetics or cladistics.
- Simulating sequences to test other methods and algorithms.
Neutral, independent, finite sites models
Most substitution models used to date are neutral, independent, finite sites models.
- Neutral
- Selection does not operate on the substitutions, and so they are unconstrained.
- Independent
- Changes in one site do not affect the probability of changes in another site.
- Finite Sites
- There are finitely many sites, and so over evolution, a single site can be changed multiple times. This means that, for example, if a character has value 0 at time 0 and at time t, it could be that no changes occurred, or that it changed to a 1 and back to a 0, or that it changed to a 1 and back to a 0 and then to a 1 and then back to a 0, and so on.
The molecular clock and the units of time
Typically, a branch length of a phylogenetic tree is expressed as the expected number of substitutions per site; if the evolutionary model indicates that each site within an ancestral sequence will typically experience x substitutions by the time it evolves to a particular descendant's sequence then the ancestor and descendant are considered to be separated by branch length x.
Sometimes a branch length is measured in terms of geological years. For example, a fossil record may make it possible to determine the number of years between an ancestral species and a descendant species. Because some species evolve at faster rates than others, these two measures of branch length are not always in direct proportion. The expected number of substitutions per site per year is often indicated with the Greek letter mu (μ).
A model is said to have a strict molecular clock if the expected number of substitutions per year μ is constant regardless of which species' evolution is being examined. An important implication of a strict molecular clock is that the number of expected substitutions between an ancestral species and any of its present-day descendants must be independent of which descendant species is examined.
Note that the assumption of a strict molecular clock is often unrealistic, especially across long periods of evolution. For example, even though rodents are genetically very similar to primates, they have undergone a much higher number of substitutions in the estimated time since divergence in some regions of the genome.[1] This could be due to their shorter generation time,[2] higher metabolic rate, increased population structuring, increased rate of speciation, or smaller body size.[3][4] When studying ancient events like the Cambrian explosion under a molecular clock assumption, poor concurrence between cladistic and phylogenetic data is often observed. There has been some work on models allowing variable rate of evolution (see for example [5] and [6]).
Models that can take into account variability of the rate of the molecular clock between different evolutionary lineages in the phylogeny are called “relaxed” in opposition to “strict”. In such models the rate can be assumed to be correlated or not between ancestors and descendants and rate variation among lineages can be drawn from many distributions but usually exponential and lognormal distributions are applied. There is a special case, called “local molecular clock” when a phylogeny is divided into at least two partitions (sets of lineages) and in each strict molecular clock is applied but with different rate.
Time-reversible and stationary models
Many useful substitution models are time-reversible; in terms of the mathematics, the model does not care which sequence is the ancestor and which is the descendant so long as all other parameters (such as the number of substitutions per site that is expected between the two sequences) are held constant.
When an analysis of real biological data is performed, there is generally no access to the sequences of ancestral species, only to the present-day species. However, when a model is time-reversible, which species was the ancestral species is irrelevant. Instead, the phylogenetic tree can be rooted using any of the species, re-rooted later based on new knowledge, or left unrooted. This is because there is no 'special' species, all species will eventually derive from one another with the same probability.
A model is time reversible if and only if it satisfies the property (the notation is explained below)
or, equivalently, the detailed balance property,
for every i, j, and t.
Time-reversibility should not be confused with stationarity. A model is stationary if Q does not change with time. The analysis below assumes a stationary model.
The mathematics of substitution models
Stationary, neutral, independent, finite sites models (assuming a constant rate of evolution) have two parameters, π, an equilibrium vector of base (or character) frequencies and a rate matrix, Q, which describes the rate at which bases of one type change into bases of another type; element 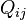 for i ≠ j is the rate at which base i goes to base j. The diagonals of the Q matrix are chosen so that the rows sum to zero:
for i ≠ j is the rate at which base i goes to base j. The diagonals of the Q matrix are chosen so that the rows sum to zero:
The equilibrium row vector π must be annihilated by the rate matrix Q:
The transition matrix function is a function from the branch lengths (in some units of time, possibly in substitutions), to a matrix of conditional probabilities. It is denoted 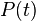 . The entry in the ith column and the jth row,
. The entry in the ith column and the jth row, 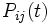 , is the probability, after time t, that there is a base j at a given position, conditional on there being a base i in that position at time 0. When the model is time reversible, this can be performed between any two sequences, even if one is not the ancestor of the other, if you know the total branch length between them.
, is the probability, after time t, that there is a base j at a given position, conditional on there being a base i in that position at time 0. When the model is time reversible, this can be performed between any two sequences, even if one is not the ancestor of the other, if you know the total branch length between them.
The asymptotic properties of Pij(t) are such that Pij(0) = δij, where δij is the Kronecker delta function. That is, there is no change in base composition between a sequence and itself. At the other extreme, 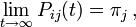 or, in other words, as time goes to infinity the probability of finding base j at a position given there was a base i at that position originally goes to the equilibrium probability that there is base j at that position, regardless of the original base. Furthermore, it follows that
or, in other words, as time goes to infinity the probability of finding base j at a position given there was a base i at that position originally goes to the equilibrium probability that there is base j at that position, regardless of the original base. Furthermore, it follows that 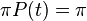 for all t.
for all t.
The transition matrix can be computed from the rate matrix via matrix exponentiation:
where Qn is the matrix Q multiplied by itself enough times to give its nth power.
If Q is diagonalizable, the matrix exponential can be computed directly: let Q = U−1 Λ U be a diagonalization of Q, with
where Λ is a diagonal matrix and where 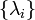 are the eigenvalues of Q, each repeated according to its multiplicity. Then
are the eigenvalues of Q, each repeated according to its multiplicity. Then
where the diagonal matrix eΛt is given by
GTR: Generalised time reversible
GTR is the most general neutral, independent, finite-sites, time-reversible model possible. It was first described in a general form by Simon Tavaré in 1986.[7]
The GTR parameters for nucleotides consist of an equilibrium base frequency vector, 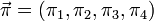 , giving the frequency at which each base occurs at each site, and the rate matrix
, giving the frequency at which each base occurs at each site, and the rate matrix
Because the model must be time reversible and must approach the equilibrium nucleotide (base) frequencies at long times, each rate below the diagonal equals the reciprocal rate above the diagonal multiplied by the equilibrium ratio of the two bases. As such, the nucleotide GTR requires 6 substitution rate parameters and 4 equilibrium base frequency parameters. Since the 4 frequency parameters must sum to 1, there are only 3 free frequency parameters. The total of 9 free parameters is often further reduced to 8 parameters plus 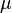 , the overall number of substitutions per unit time. When measuring time in substitutions (
, the overall number of substitutions per unit time. When measuring time in substitutions (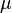 =1) only 8 free parameters remain.
=1) only 8 free parameters remain.
In general, to compute the number of parameters, you count the number of entries above the diagonal in the matrix, i.e. for n trait values per site 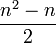 , and then add n-1 for the equilibrium frequencies, and subtract 1 because
, and then add n-1 for the equilibrium frequencies, and subtract 1 because 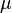 is fixed. You get
is fixed. You get
For example, for an amino acid sequence (there are 20 "standard" amino acids that make up proteins), you would find there are 208 parameters. However, when studying coding regions of the genome, it is more common to work with a codon substitution model (a codon is three bases and codes for one amino acid in a protein). There are 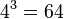 codons, resulting in 2078 free parameters, but when the rates for transitions between codons which differ by more than one base are assumed to be zero, then there are only
codons, resulting in 2078 free parameters, but when the rates for transitions between codons which differ by more than one base are assumed to be zero, then there are only 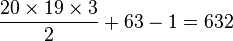 parameters.
parameters.
Mechanistic vs. empirical models
A main difference in evolutionary models is how many parameters are estimated every time for the data set under consideration and how many of them are estimated once on a large data set. Mechanistic models describe all substitutions as a function of a number of parameters which are estimated for every data set analyzed, preferably using maximum likelihood. This has the advantage that the model can be adjusted to the particularities of a specific data set (e.g. different composition biases in DNA). Problems can arise when too many parameters are used, particularly if they can compensate for each other. Then it is often the case that the data set is too small to yield enough information to estimate all parameters accurately.
Empirical models are created by estimating many parameters (typically all entries of the rate matrix and the character frequencies, see the GTR model above) from a large data set. These parameters are then fixed and will be reused for every data set. This has the advantage that those parameters can be estimated more accurately. Normally, it is not possible to estimate all entries of the substitution matrix from the current data set only. On the downside, the estimated parameters might be too generic and do not fit a particular data set well enough.
With the large-scale genome sequencing still producing very large amounts of DNA and protein sequences, there is enough data available to create empirical models with any number of parameters. Because of the problems mentioned above, the two approaches are often combined, by estimating most of the parameters once on large-scale data, while a few remaining parameters are then adjusted to the data set under consideration. The following sections give an overview of the different approaches taken for DNA, protein or codon-based models.
Models of DNA substitution
See main article: Models of DNA evolution for more formal descriptions of the DNA models.
Models of DNA evolution were first proposed in 1969 by Jukes and Cantor,[8] assuming equal transition rates as well as equal equilibrium frequencies for all bases. In 1980 Kimura[9] introduced a model with two parameters: one for the transition and one for the transversion rate and in 1981, Felsenstein[10] proposed a four-parameter model in which the substitution rate corresponds to the equilibrium frequency of the target nucleotide. Hasegawa, Kishino and Yano (HKY)[11] unified the two last models to a five-parameter model. In the 1990s, models similar to HKY were developed and refined by several researchers.[12][13][14]
For DNA substitution models, mainly mechanistic models (as described above) are employed. The small number of parameters to estimate makes this feasible, but also DNA is often highly optimized for specific purposes (e.g. fast expression or stability) depending on the organism and the type of gene, making it necessary to adjust the model to these circumstances.
Models of amino acid substitutions
For many analyses, particularly for longer evolutionary distances, the evolution is modeled on the amino acid level. Since not all DNA substitution also alter the encoded amino acid, information is lost when looking at amino acids instead of nucleotide bases. However, several advantages speak in favor of using the amino acid information: DNA is much more inclined to show compositional bias than amino acids, not all positions in the DNA evolve at the same speed (non-synonymous mutations are more likely to become fixed in the population than synonymous ones), but probably most important, because of those fast evolving positions and the limited alphabet size (only four possible states), the DNA suffers much more from back substitutions, making it difficult to accurately estimate longer distances.
Unlike the DNA models, amino acid models traditionally are empirical models. They were pioneered in the 1970s by Dayhoff and co-workers ,[15] by estimating replacement rates from protein alignments with at least 85% identity. This minimized the chances of observing multiple substitutions at a site. From the estimated rate matrix, a series of replacement probability matrices were derived, known under names such as PAM250. The Dayhoff model was used to assess the significance of homology search results, but also for phylogenetic analyses. The Dayhoff PAM matrices were based on relatively few alignments (since not more were available at that time), but in the 1990s, new matrices were estimated using almost the same methodology, but based on the large protein databases available then (,[16][17] the latter being known as "JTT" matrices).
References
- ↑ Gu X, Li WH (September 1992). "Higher rates of amino acid substitution in rodents than in humans". Mol. Phylogenet. Evol. 1 (3): 211–4. doi:10.1016/1055-7903(92)90017-B. PMID 1342937.
- ↑ Li WH, Ellsworth DL, Krushkal J, Chang BH, Hewett-Emmett D (February 1996). "Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis". Mol. Phylogenet. Evol. 5 (1): 182–7. doi:10.1006/mpev.1996.0012. PMID 8673286.
- ↑ Martin AP, Palumbi SR (May 1993). "Body size, metabolic rate, generation time, and the molecular clock". Proc. Natl. Acad. Sci. U.S.A. 90 (9): 4087–91. doi:10.1073/pnas.90.9.4087. PMC 46451. PMID 8483925.
- ↑ Yang Z, Nielsen R (April 1998). "Synonymous and nonsynonymous rate variation in nuclear genes of mammals". J. Mol. Evol. 46 (4): 409–18. doi:10.1007/PL00006320. PMID 9541535.
- ↑ Kishino H, Thorne JL, Bruno WJ (March 2001). "Performance of a divergence time estimation method under a probabilistic model of rate evolution". Mol. Biol. Evol. 18 (3): 352–61. doi:10.1093/oxfordjournals.molbev.a003811. PMID 11230536.
- ↑ Thorne JL, Kishino H, Painter IS (December 1998). "Estimating the rate of evolution of the rate of molecular evolution". Mol. Biol. Evol. 15 (12): 1647–57. doi:10.1093/oxfordjournals.molbev.a025892. PMID 9866200.
- ↑ Tavaré S. "Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences". Lectures on Mathematics in the Life Sciences (American Mathematical Society) 17: 57–86.
- ↑ Jukes, T.H., Cantor, C.R. (1969). "Evolution of protein molecules". In Munro, H.N. Mammalian protein metabolism. New York: Academic Press. pp. 21–123.
- ↑ Kimura M (December 1980). "A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences". J. Mol. Evol. 16 (2): 111–20. doi:10.1007/BF01731581. PMID 7463489.
- ↑ Felsenstein J (1981). "Evolutionary trees from DNA sequences: a maximum likelihood approach". J. Mol. Evol. 17 (6): 368–76. doi:10.1007/BF01734359. PMID 7288891.
- ↑ Hasegawa M, Kishino H, Yano T (1985). "Dating of the human-ape splitting by a molecular clock of mitochondrial DNA". J. Mol. Evol. 22 (2): 160–74. doi:10.1007/BF02101694. PMID 3934395.
- ↑ Tamura K (July 1992). "Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases". Mol. Biol. Evol. 9 (4): 678–87. PMID 1630306.
- ↑ Tamura K, Nei M (May 1993). "Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees". Mol. Biol. Evol. 10 (3): 512–26. PMID 8336541.
- ↑ Halpern, AL; Bruno, WJ (July 1998). "Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies". Mol. Biol. Evol. 15 (7): 910–7. doi:10.1093/oxfordjournals.molbev.a025995. PMID 9656490.
- ↑ Dayhoff MO, Schwartz RM, Orcutt BC (1978). "A model for evolutionary change in proteins". Atlas of Protein Sequence and Structure 5: 345–352.
- ↑ Gonnet GH, Cohen MA, Benner SA (1992). "Exhaustive matching of the entire protein sequence database". Science 256 (5062): 1443–5. doi:10.1126/science.1604319. PMID 1604319.
- ↑ Jones DT, Taylor WR, Thornton JM (1992). "The rapid generation of mutation data matrices from protein sequences". Comput Applic Biosci 8: 275–282. doi:10.1093/bioinformatics/8.3.275.