Structure validation
Macromolecular structure validation is the process of evaluating reliability for 3-dimensional atomic models of large biological molecules such as proteins and nucleic acids. These models, which provide 3D coordinates for each atom in the molecule (see example in the image), come from structural biology experiments such as x-ray crystallography[1] or nuclear magnetic resonance (NMR).[2] The validation has three aspects: 1) checking on the validity of the thousands to millions of measurements in the experiment; 2) checking how consistent the atomic model is with those experimental data; and 3) checking consistency of the model with known physical and chemical properties.
Proteins and nucleic acids are the workhorses of biology, providing the necessary chemical reactions, structural organization, growth, mobility, reproduction, and environmental sensitivity. Essential to their biological functions are the detailed 3D structures of the molecules and the changes in those structures. To understand and control those functions, we need accurate knowledge about the models that represent those structures, including their many strong points and their occasional weaknesses.
End-users of macromolecular models include clinicians, teachers and students, as well as the structural biologists themselves, journal editors and referees, experimentalists studying the macromolecules by other techniques, and theoreticians and bioinformaticians studying more general properties of biological molecules. Their interests and requirements vary, but all benefit greatly from a global and local understanding of the reliability of the models.
Historical summary
Macromolecular crystallography was preceded by the older field of small-molecule x-ray crystallography (for structures with less than a few hundred atoms). Small-molecule diffraction data extends to much higher resolution than feasible for macromolecules, and has a very clean mathematical relationship between the data and the atomic model. The residual, or R-factor, measures the agreement between the experimental data and the values back-calculated from the atomic model. For a well-determined small-molecule structure the R-factor is nearly as small as the uncertainty in the experimental data (well under 5%). Therefore that one test by itself provides most of the validation needed, but a number of additional consistency and methodology checks are done by automated software[3] as a requirement for small-molecule crystal structure papers submitted to the International Union of Crystallography (IUCr) journals such as Acta Crystallographica section B or C. Atomic coordinates of these small-molecule structures are archived and accessed through the Cambridge Structural Database (CSD)[4] or the Crystallography Open Database (COD).[5]
The first macromolecular validation software was developed around 1990, for proteins. It included Rfree cross-validation for model-to-data match,[6] bond length and angle parameters for covalent geometry,[7] and sidechain and backbone conformational criteria.[8][9][10] For macromolecular structures, the atomic models are deposited in the Protein Data Bank (PDB), still the single archive of this data. The PDB was established in the 1970s at Brookhaven National Laboratory,[11] moved in 2000 to the (Research Collaboration for Structural Biology) centered at Rutgers,[12] and expanded in 2003 to become the wwPDB (worldwide Protein Data Bank),[13] with access sites added in Europe () and Asia (), and with NMR data handled at the BioMagResBank (BMRB) in Wisconsin.
Validation rapidly became standard in the field,[14] with further developments described below. *Obviously needs expansion*
A large boost was given to the applicability of comprehensive validation for both x-ray and NMR as of February 1, 2008, when the worldwide Protein Data Bank (wwPDB) made mandatory the deposition of experimental data along with atomic coordinates. Since 2012 strong forms of validation have been in the process of being adopted for wwPDB deposition from recommendations of the wwPDB Validation Task Force committees for x-ray crystallography,[15] for NMR,[16] for SAXS (small-angle x-ray scattering), and for cryoEM (cryo-Electron Microscopy).[17]
Validation for Crystallography (X-ray and Neutron)
Overall Considerations
Global vs Local Criteria
Many evaluation criteria apply globally to an entire experimental structure, most notably the resolution, the anisotropy or incompleteness of the data, and the residual or R-factor that measures overall model-to-data match (see below). Those help a user choose the most accurate among related Protein Data Bank entries to answer their questions. Other criteria apply to individual residues or local regions in the 3D structure, such as fit to the local electron density map or steric clashes between atoms. Those are especially valuable to the structural biologist for making improvements to the model, and to the user for evaluating the reliability of that model right around the place they care about - such as a site of enzyme activity or drug binding. Both types of measures are very useful, but although global criteria are easier to state or publish, local criteria make the greatest contribution to scientific accuracy and biological relevance. As expressed in the Rupp textbook, "Only local validation, including assessment of both geometry and electron density, can give an accurate picture of the reliability of the structure model or any hypothesis based on local features of the model."[18]
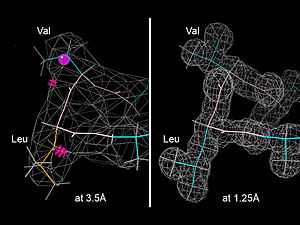
Model Validation
Geometry
Conformation (Dihedrals): Protein & RNA
The backbone and sidechain dihedral angles of protein and RNA have been shown to have specific combinations of angles which are allowed.
Software and Websites
- rcsbPDB validation/deposition site
- MolProbity web service
- Protein structure validation database PDBREPORT.
- EDS (Electron Density Server)[21]
- What_Check software
- ProCheck software
- Coot modeling software (built-in validation)[22]
- OOPS2, part of the Uppsala Software Factory
- ProSA web service
- Verify-3D profile analysis
- PDB_REDO optimized X-ray structure models[23]
- PROSESS - Protein Structure Evaluation Suite & Server
- Resolution by Proxy, ResProx - protein model resolution-by-proxy
- VADAR - Volume, Area, Dihedral Angle Reporter
For NMR (Nuclear Magnetic Resonance)
Data Validation: Chemical Shifts, NOEs, RDCs
AVS. Assignment validation suite (AVS) checks the chemical shifts list in BioMagResBank (BMRB) format for problems.
PSVS (Protein Structure Validation Server at the NESG)[24]
PROSESS. PROSESS (Protein Structure Evaluation Suite & Server) is a new web server that offers an assessment of protein structural models by NMR chemical shifts as well as NOEs, geometrical, and knowledge-based parameters.
LACS. Linear analysis of chemical shifts is used for absolute referencing of chemical shift data.
Model-to-Data Validation
TALOS+. Predicts protein backbone torsion angles from chemical shift data. Frequently used to generate further restraints applied to a structure model during refinement.
Model Validation: As Above

Dynamics: Core vs Loops, Tails, and Mobile Domains
One of the critical needs for NMR structural ensemble validation is to distinguish well-determined regions (those that have experimental data) from regions that are highly mobile and/or have no observed data. There are several current or proposed methods for making this distinction such as Random Coil Index, but so far the NMR community has not standardized on one.
Software and Websites
- PSVS (Protein Structure Validation Server at the NESG)[24]
- CING (Common Interface for NMR structure Generation) software
- ProCheckNMR software[25]
- TALOS+ Software & Server(server for predicting protein backbone torsion angles from chemical shift)
- VADAR - Volume, Area, Dihedral Angle Reporter
- PROSESS - Protein Structure Evaluation Suite & Server
- ResProx - protein model resolution-by-proxy
- MolProbity (includes analyses for NMR)
For Cryo-Electron Microscopy and Hybrid Methods
Software and Websites
For SAXS (Small-Angle X-ray Scattering)
SAXS is a rapidly growing area of structure determination, both as a source of approximate 3D structure for initial or difficult cases and as a component of hybrid-method structure determination when combined with NMR, EM, crystallographic, cross-linking, or computational information. There is great interest in the development of reliable validation standards for SAXS data interpretation and for quality of the resulting models, but there are as yet no established methods in general use. Three recent steps in this direction are the creation of a Small-Angle Scattering Validation Task Force committee by the worldwide Protein DataBank and its initial report,[26] a set of suggested standards for data inclusion in publications,[27] and an initial proposal of statistically derived criteria for automated quality evaluation.[28]
For Computational Biology
It is difficult to do meaningful validation of an individual, purely computational, macromolecular model in the absence of experimental data for that molecule, because the model with the best geometry and conformational score may not be the one closest to the right answer. Therefore, much of the emphasis in validation of computational modeling is in assessment of the methods. To avoid bias and wishful thinking, double-blind prediction competitions have been organized, the original example of which (held every 2 years since 1994) is CASP (Critical Assessment of Structure Prediction) to evaluate predictions of 3D protein structure for newly solved crystallographic or NMR structures held in confidence until the end of the relevant competition.[29] The major critierion for CASP evaluation is a weighted score called GDT-TS for the match of Calpha positions between the predicted and the experimental models.[30]
Software and Websites
References
- ↑ Rupp 2009
- ↑ Cavanagh 2006
- ↑ Spek AL (2003). "Single-crystal structure validation with the program PLATON". J. Applied Crystallography 36: 7–13. doi:10.1107/S0021889802022112.
- ↑ Allen FH (2002). "The Cambridge Structural Database: a quarter of a million crystal structures and rising". Acta Crystallographica B 58: 380–388. doi:10.1107/S0108768102003890.
- ↑ Grazulis S, Chateigner D, Downs RT, Yokochi AT, Quiros M, Lutterotti L, Manakova E, Butkus J, Moeck, P, Le Bail A (2009). "Crystallography Open Database – an open-access collection of crystal structures". Journal of Applied Crystallography 42: 726–729. doi:10.1107/s0021889809016690.
- ↑ Brunger AT (1992). "Free R value: a novel statistical quantity for assessing the accuracy of crystal structures". Nature (London) 355 (6359): 472–475. doi:10.1038/355472a0. PMID 18481394.
- ↑ 7.0 7.1 Engh RA, Huber R (1991). "Accurate bond and angle parameters for X-ray protein structure refinement". Acta Crystallographica A 47: 392–400. doi:10.1107/s0108767391001071.
- ↑ Ponder JW, Richards FM (1987). "Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes". J. Molecular Biology 193: 775–791.
- ↑ Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993). "PROCHECK: a program to check the stereochemical quality of protein structures". J. Applied Crystallography 26: 283–291. doi:10.1107/s0021889892009944.
- ↑ Hooft RWW, Vriend G, Sander C, Abola EE (1996). "Errors in protein structures". Nature (London) 381 (6580): 272. doi:10.1038/381272a0. PMID 8692262.
- ↑ Bernstein FC, Koetzle TF, Williams GJB, Meyer EF Jr, Brice MD, Rodgers JR, Kennard 0, Shimanouchi T, Tasumi M (1977). "The Protein Data Bank: A computer-based archival file for macromolecular structures". J. Molecular Biology 112 (3): 535–542. doi:10.1016/s0022-2836(77)80200-3. PMID 875032.
- ↑ Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000). "The Protein Data Bank". Nucleic Acids Research 28 (1): 235–242. doi:10.1093/nar/28.1.235. PMC 102472. PMID 10592235.
- ↑ Berman HM, Henrick K, Nakamura H (2003). "Announcing the worldwide Protein Data Bank". Nature Structural & Molecular Biology 10 (12): 980. doi:10.1038/nsb1203-980. PMID 14634627.
- ↑ Kleywegt GJ (2000). "Validation of protein crystal structures". Acta Crystallogr D 56: 18–19.
- ↑ Read RJ, Adams PD, Arendall WB III, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lutteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH (2011). "A New Generation of Crystallographic Validation Tools for the Protein Data Bank". Structure 19 (10): 1395–1412. doi:10.1016/j.str.2011.08.006. PMC 3195755. PMID 22000512.
- ↑ Montelione GT, Nilges M, Bax A, Guntert P, Herrmann T, Richardson JS, Schwieters C, Vranken WF, Vuister GW, Wishart DS, Berman HM, Kleywegt GJ, Markley JL (2013). "Recommendations of the NMR Structure Validation Task Force". Structure 21 (9): 1563–1570. doi:10.1016/j.str.2013.07.021. PMC 3884077. PMID 24010715.
- ↑ Henderson R, Sali A, Baker ML, Carragher B, Devkota B, Downing KH, Egelman EH, Feng Z, Frank J, Grigorieff N, Jiang W, Ludtke SJ, Medalia O, Penczek PA, Rosenthal PB, Rossmann MG, Schmid MF, Schroeder GF, Steven AC, Stokes, DL, Westbrook JD, Wriggers W, Yang H, Young J, Berman HM, Chiu W, Kleywegt GJ, Lawson CL (2012). "Outcome of the First Electron Microscopy Validation Task Force Meeting". Structure 20: 205–214. doi:10.1016/j.str.2011.12.014.
- ↑ Rupp 2009, Chapter 13, Key Concepts
- ↑ Gelbin A, Schneider B, Clowney L, Hsieh S-H, Olson WK, Berman HM (1996). "Geometric parameters in Nucleic Acids:Sugar and Phosphate Constituents". J Amer Chem Soc 118: 519–529. doi:10.1021/ja9528846.
- ↑ Schultze P, Feigon J (1997). "Chirality errors in nucleic acid structures". Nature (London) 387: 668.
- ↑ Kleywegt G.J., Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA (2004). "The Uppsala Electron-Density Server". Acta Crystallographica D 60: 2240–2249. doi:10.1107/s0907444904013253.
- ↑ Emsley P, Lohkamp B, Scott WG, Cowtan K (2010). "Features and development of Coot". Acta Crystallographica D 66: 486–501. doi:10.1107/s0907444910007493.
- ↑ Joosten RP, Joosten K, Murshudov GN, Perrakis A (2012). "PDB_REDO: constructive validation, more than just looking for errors". Acta Crystallographica D 68: 484–496. doi:10.1107/s0907444911054515.
- ↑ 24.0 24.1 Huang YJ, Powers R and Montelione GT (2005). "Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics". Journal of Biomolecular NMR 127 (6): 1665–1674. doi:10.1021/ja047109h. PMID 15701001.
- ↑ Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996). "AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR". Journal of the American Chemical Society 8 (4): 477–486. doi:10.1007/bf00228148. PMID 9008363.
- ↑ Trewhella J, Hendrickson WA, Kleywegt GJ, Sali A, Sato M, Schwede T, Svergun DI, Tainer JA, Westbrook J, Berman HM (2013). "Report of the wwPDB Small-Angle Scattering Task Force: Data requirements for biomolecular modeling and the PDB structure". Structure 21: 875–881. doi:10.1016/j.str.2013.04.020.
- ↑ Jacques DA, Guss JM, Svergun DI, Trewhella J (2012). "Publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution". Acta Crystallographica D 68: 620–626. doi:10.1107/S0907444912012073.
- ↑ Grant TD, Luft JR, Carter LG, Matsui T, Weiss ™, Martel A, Snell EH (2015). "The accurate assessment of small-angle X-ray scattering data". Acta Crystallographica D 71: 45–56. doi:10.1107/S1399004714010876.
- ↑ Moult, J., et al. (1995). "A large-scale experiment to assess protein structure prediction methods". Proteins 23 (3): ii–iv. doi:10.1002/prot.340230303.
- ↑ Zemla, A. (2003). "LGA: a method for finding 3D similarities in protein structures". Nucleic Acids Research 31 (13): 3370–4. doi:10.1093/nar/gkg571. PMC 168977. PMID 12824330.
Further reading
- Cavanagh, John, Fairbrother, Wayne J., Palmer, Arthur G. III, Skelton, Nicholas J. (2006). Protein NMR Spectroscopy: Principles and Practice (2nd ed.). Academic Press. ISBN 0-12-164491-X.
- Rupp, Bernhard (2009). Biomolecular Crystallography: Principles, Practice, and Application to Structural Biology. Garland Science. ISBN 0815340818.