Staphylococcus aureus
| Staphylococcus aureus | |
|---|---|
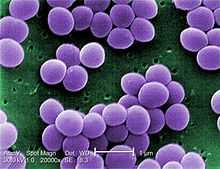 | |
| Scanning electron micrograph of S. aureus; false color added. | |
| Scientific classification | |
| Domain: | Bacteria |
| Kingdom: | Eubacteria |
| Phylum: | Firmicutes |
| Class: | Coccus |
| Order: | Bacillales |
| Family: | Staphylococcaceae |
| Genus: | Staphylococcus |
| Species: | S. aureus |
| Binomial name | |
| Staphylococcus aureus Rosenbach 1884 | |
Staphylococcus aureus is a gram-positive coccal bacterium that is a member of the Firmicutes, and is frequently found in the human respiratory tract and on the skin. It is positive for catalase and nitrate reduction. Although S. aureus is not always pathogenic, it is a common cause of skin infections (e.g. boils), respiratory disease (e.g. sinusitis), and food poisoning. Disease-associated strains often promote infections by producing potent protein toxins, and expressing cell-surface proteins that bind and inactivate antibodies. The emergence of antibiotic-resistant forms of pathogenic S. aureus (e.g. MRSA) is a worldwide problem in clinical medicine.
Staphylococcus was first identified in 1880 in Aberdeen, Scotland, by the surgeon Sir Alexander Ogston in pus from a surgical abscess in a knee joint.[1] This name was later appended to Staphylococcus aureus by Friedrich Julius Rosenbach who was credited by the official system of nomenclature at the time. It is estimated that 20% of the human population are long-term carriers of S. aureus[2] which can be found as part of the normal skin flora and in anterior nares of the nasal passages.[2][3] S. aureus is the most common species of staphylococcus to cause Staph infections and is a successful pathogen due to a combination of nasal carriage and bacterial immuno-evasive strategies.[2][3] S. aureus can cause a range of illnesses, from minor skin infections, such as pimples, impetigo, boils (furuncles), cellulitis folliculitis, carbuncles, scalded skin syndrome, and abscesses, to life-threatening diseases such as pneumonia, meningitis, osteomyelitis, endocarditis, toxic shock syndrome (TSS), bacteremia, and sepsis. Its incidence ranges from skin, soft tissue, respiratory, bone, joint, endovascular to wound infections. It is still one of the five most common causes of nosocomial infections and is often the cause of postsurgical wound infections. Each year, some 500,000 patients in United States' hospitals contract a staphylococcal infection.[4]
| Staphylococcus aureus | |
|---|---|
| Classification and external resources | |
| ICD-9 | 041.11 |
Microbiology


S. aureus (/ˌstæfɨlɵˈkɒkəs ˈɔriəs/, Greek σταφυλόκοκκος, "grape-cluster berry", Latin aureus, "golden") is a facultative anaerobic gram-positive coccal bacterium also known as "golden staph" and Oro staphira. In medical literature the bacteria is often referred to as S. aureus or Staph aureus. Staphylococcus should not be confused with the similarly named and medically relevant genus Streptococcus. S. aureus appears as grape-like clusters when viewed through a microscope, and has large, round, golden-yellow colonies, often with hemolysis, when grown on blood agar plates.[5] S. aureus reproduces asexually by binary fission. The two daughter cells do not fully separate and remain attached to one another. This is why the cells are observed in clusters.[6]
S. aureus is catalase-positive (meaning it can produce the enzyme catalase). Catalase converts hydrogen peroxide (H
2O
2) to water and oxygen. Catalase-activity tests are sometimes used to distinguish staphylococci from enterococci and streptococci. Previously, S. aureus was differentiated from other staphylococci by the coagulase test. However it is now known that not all S. aureus are coagulase-positive[5][7] and that incorrect species identification can impact effective treatment and control measures.[8]
Role in disease

S. aureus is responsible for many infections but it may also occur as a commensal. The presence of S. aureus does not always indicate infection. S. aureus can survive from hours to weeks, or even months, on dry environmental surfaces, depending on strain.[9]
S. aureus can infect tissues when the skin or mucosal barriers have been breached. This can lead to many different types of infections including furuncles and carbuncles (a collection of furuncles).
S. aureus infections can spread through contact with pus from an infected wound, skin-to-skin contact with an infected person by producing hyaluronidase that destroys tissues, and contact with objects such as towels, sheets, clothing, or athletic equipment used by an infected person. Deeply penetrating S. aureus infections can be severe. Prosthetic joints put a person at particular risk of septic arthritis, and staphylococcal endocarditis (infection of the heart valves) and pneumonia. Strains of S. aureus can host phages, such as Φ-PVL (produces Panton-Valentine leukocidin), that increase virulence.
Atopic dermatitis
S. aureus is extremely prevalent in persons with atopic dermatitis. It is mostly found in fertile, active places, including the armpits, hair, and scalp. Large pimples that appear in those areas may exacerbate the infection if lacerated. This can lead to staphylococcal scalded skin syndrome (SSSS). A severe form of this, Ritter's disease, can be observed in neonates.[10]
The presence of S. aureus in persons with atopic dermatitis is not an indication to treat with oral antibiotics, as evidence has not shown this to give benefit to the patient.[11] The relationship between S. aureus and atopic dermatitis is unclear.[11] Evidence shows that attempting to control S. aureus with oral antibiotics is not efficacious.[11]
Animal infections
S. aureus can survive on dogs,[12] cats,[13] and horses,[14] and can cause bumblefoot in chickens.[15] Some believe health-care workers' dogs should be considered a significant source of antibiotic-resistant S. aureus, especially in times of outbreak.[12] S. aureus is one of the causal agents of mastitis in dairy cows. Its large polysaccharide capsule protects the organism from recognition by the cow's immune defenses.[16]
Virulence factors
Enzymes
Staphylococcus aureus produces various enzymes such as coagulase (bound and free coagulases) which clots plasma and coats the bacterial cell to probably prevent phagocytosis. Hyaluronidase (also known as spreading factor) breaks down hyaluronic acid and helps in spreading of Staphylococcus aureus. S.aureus also produces DNAse (deoxyribonuclease) which breaks down the DNA, lipase to digest lipids, staphylokinase to dissolve fibrin and aid in spread, and beta-lactamase for drug resistance.[17]
Toxins
Depending on the strain, S. aureus is capable of secreting several exotoxins, which can be categorized into three groups. Many of these toxins are associated with specific diseases.[18]
- Superantigens
- (PTSAgs) have superantigen activities that induce toxic shock syndrome (TSS). This group includes the toxin TSST-1, enterotoxin type B, which causes TSS associated with tampon use. This is characterized by fever, erythematous rash, hypotension, shock, multiple organ failure, and skin desquamation. Lack of antibody to TSST-1 plays a part in the pathogenesis of toxic shock syndrome. Other strains of S. aureus can produce an enterotoxin that is the causative agent of S. aureus gastroenteritis. This gastroenteritis is self-limiting, characterized by vomiting and diarrhea one to six hours after ingestion of the toxin with recovery in eight to 24 hours. Symptoms include nausea, vomiting, diarrhea, and major abdominal pain.[19][20]
- Exfoliative toxins
- EF toxins are implicated in the disease staphylococcal scalded-skin syndrome (SSSS), which occurs most commonly in infants and young children. It also may occur as epidemics in hospital nurseries. The protease activity of the exfoliative toxins causes peeling of the skin observed with SSSS.[20]
- Other toxins
- Staphylococcal toxins that act on cell membranes include alpha toxin, beta toxin, delta toxin, and several bicomponent toxins. The bicomponent toxin Panton-Valentine leukocidin (PVL) is associated with severe necrotizing pneumonia in children.[21][22] The genes encoding the components of PVL are encoded on a bacteriophage found in community-associated methicillin-resistant S. aureus (MRSA) strains.
Other immunoevasive strategies
- Protein A
Protein A is anchored to staphylococcal peptidoglycan pentaglycine bridges (chains of five glycine residues) by the transpeptidase sortase A.[23] Protein A, an IgG-binding protein, binds to the Fc region of an antibody. In fact, studies involving mutation of genes coding for protein A resulted in a lowered virulence of S. aureus as measured by survival in blood, which has led to speculation that protein A-contributed virulence requires binding of antibody Fc regions.[24]
Protein A in various recombinant forms has been used for decades to bind and purify a wide range of antibodies by immunoaffinity chromatography. Transpeptidases, such as the sortases responsible for anchoring factors like Protein A to the staphylococcal peptidoglycan, are being studied in hopes of developing new antibiotics to target MRSA infections.[25]

- Staphylococcal pigments
Some strains of S. aureus are capable of producing staphyloxanthin — a golden coloured carotenoid pigment. This pigment acts as a virulence factor, primarily by being a bacterial antioxidant which helps the microbe evade the reactive oxygen species which the host immune system uses to kill pathogens.[26][27]
Mutant strains of S. aureus modified to lack staphyloxanthin are less likely to survive incubation with an oxidizing chemical, such as hydrogen peroxide than pigmented strains. Mutant colonies are quickly killed when exposed to human neutrophils, while many of the pigmented colonies survive.[26] In mice, the pigmented strains cause lingering abscesses when inoculated into wounds, whereas wounds infected with the unpigmented strains quickly heal.
These tests suggest the Staphylococcus strains use staphyloxanthin as a defence against the normal human immune system. Drugs designed to inhibit the production of staphyloxanthin may weaken the bacterium and renew its susceptibility to antibiotics.[27] In fact, because of similarities in the pathways for biosynthesis of staphyloxanthin and human cholesterol, a drug developed in the context of cholesterol-lowering therapy was shown to block S. aureus pigmentation and disease progression in a mouse infection model.[28]
Classical diagnosis
Depending upon the type of infection present, an appropriate specimen is obtained accordingly and sent to the laboratory for definitive identification by using biochemical or enzyme-based tests. A Gram stain is first performed to guide the way, which should show typical gram-positive bacteria, cocci, in clusters. Second, the isolate is cultured on mannitol salt agar, which is a selective medium with 7–9% NaCl that allows S. aureus to grow, producing yellow-colored colonies as a result of mannitol fermentation and subsequent drop in the medium's pH.
Furthermore, for differentiation on the species level, catalase (positive for all Staphylococcus species), coagulase (fibrin clot formation, positive for S. aureus), DNAse (zone of clearance on DNase agar), lipase (a yellow color and rancid odor smell), and phosphatase (a pink color) tests are all done. For staphylococcal food poisoning, phage typing can be performed to determine whether the staphylococci recovered from the food were the source of infection.
Rapid diagnosis and typing
Diagnostic microbiology laboratories and reference laboratories are key for identifying outbreaks and new strains of S. aureus. Recent genetic advances have enabled reliable and rapid techniques for the identification and characterization of clinical isolates of S. aureus in real time. These tools support infection control strategies to limit bacterial spread and ensure the appropriate use of antibiotics. Quantitative PCR is being increasingly employed in clinical laboratories as a technique to identifying outbreaks.[29][30]
When observing the evolvement of S. aureus and its ability to adapt to each modified antibiotic, in general, there are two basic methods known as “band-based” or “sequence-based”.[31] Keeping these two methods in mind, other methods like multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), bacteriophage typing, spa locus typing, and SCCmec typing are often conducted more than others.[32] With these methods,not only are we able to determine where strains of MRSA originated from, but where they currently reside.[33]
With MLST, this technique of typing utilizes fragments of several housekeeping genes known as aroE, glpF, gmk, pta, tip, and yqiL. These sequences are then assigned a number which give to a strong of several numbers that serve as the allelic profile. Although this is a common method, a limitation about this method is the maintenance of the micro-array which detects newly allelic profiles, making it a costly and time consuming experiment.[31]
With PFGE, a method which is still very much utilized dating back to its first success in 1980s remains capable of helping differentiate Methicillin-resistant S. aureus isolates.[33] To accomplish this, the technique uses multiple gel electrophoresis, along with a voltage gradient to display clear resolutions of molecules. The S. aureus fragments then transition down the gel producing specific band patters that are later compare with other isolates in hopes of identifying related strains. Limitations of the method include, practical difficulties with uniform band patterns and PFGE sensitivity as a whole.
Spa locus typing is also considered a popular technique that uses a single locus zone in a polymorphic region of S. aureus to distinguish any form of mutations.[33] Although this technique is often inexpensive and less time consuming, the chance of losing discriminatory power makes it hard to differentiate between MLST CC’s exemplifies a crucial limitation.
Treatment and antibiotic resistance
The treatment of choice for S. aureus infection is penicillin. Penicillin, an antibiotic derived from Penicillum fungus, inhibits the formation of peptidoglycan cross-linkages that provide the rigidity and strength in a bacterial cell wall. The four-membered β-lactam ring of penicillin is bound to enzyme DD-transpeptidase, an enzyme that when functional, cross-links chains of peptidoglycan that form bacterial cell walls. The binding of β-lactam to DD-transpeptidase inhibits the enzyme’s functionality and it can no longer catalyze the formation of the cross-links. As a result, cell wall formation and degradation is imbalanced, thus resulting in cell death. In most countries, however, penicillin resistance is extremely common, and first-line therapy is most commonly a penicillinase-resistant β-lactam antibiotic (for example, oxacillin or flucloxacillin, both of which have the same mechanism of action as penicillin). Combination therapy with gentamicin may be used to treat serious infections, such as endocarditis,[34][35] but its use is controversial because of the high risk of damage to the kidneys.[36] The duration of treatment depends on the site of infection and on severity.
Antibiotic resistance in S. aureus was uncommon when penicillin was first introduced in 1943. Indeed, the original petri dish on which Alexander Fleming of Imperial College London observed the antibacterial activity of the Penicillium fungus was growing a culture of S. aureus. By 1950, 40% of hospital S. aureus isolates were penicillin-resistant; and, by 1960, this had risen to 80%.[37]
Methicillin-resistant S. aureus, abbreviated MRSA and often pronounced /ˈmɜrsə/ or /ɛm ɑː ɛs eɪ/, is one of a number of greatly feared strains of S. aureus which have become resistant to most β-lactam antibiotics. For this reason, vancomycin, a glycopeptide antibiotic, is commonly used to combat MRSA. Vancomycin inhibits the synthesis of peptidoglycan, but unlike β-lactam antibiotics, glycopeptide antibiotics target and bind to amino acids in the cell wall, preventing peptidoglycan cross-linkages from forming. MRSA strains are most often found associated with institutions such as hospitals, but are becoming increasingly prevalent in community-acquired infections. A recent study by the Translational Genomics Research Institute showed that nearly half (47%) of the meat and poultry in U.S. grocery stores were contaminated with S. aureus, with more than half (52%) of those bacteria resistant to antibiotics.[38] This resistance is commonly caused by the widespread use of antibiotics in the husbandry of livestock, including prevention or treatment of an infection as well as promoting growth.
| Wikinews has related news: Supergerm deaths soar, surpass AIDS in the United States |
Researchers from Italy have identified a bacteriophage active against S. aureus, including methicillin-resistant strains (MRSA), in mice and possibly humans.[39]
Mechanisms of antibiotic resistance

Staphylococcal resistance to penicillin is mediated by penicillinase (a form of β-lactamase) production: an enzyme that cleaves the β-lactam ring of the penicillin molecule, rendering the antibiotic ineffective. Penicillinase-resistant β-lactam antibiotics, such as methicillin, nafcillin, oxacillin, cloxacillin, dicloxacillin, and flucloxacillin, are able to resist degradation by staphylococcal penicillinase.
Resistance to methicillin is mediated via the mec operon, part of the staphylococcal cassette chromosome mec (SCCmec). Resistance is conferred by the mecA gene, which codes for an altered penicillin-binding protein (PBP2a or PBP2') that has a lower affinity for binding β-lactams (penicillins, cephalosporins, and carbapenems). This allows for resistance to all β-lactam antibiotics, and obviates their clinical use during MRSA infections. As such, the glycopeptide vancomycin is often deployed against MRSA.
Aminoglycoside antibiotics, such as kanamycin, gentamicin, streptomycin, etc., were once effective against staphylococcal infections until strains evolved mechanisms to inhibit the aminoglycosides' action, which occurs via protonated amine and/or hydroxyl interactions with the ribosomal RNA of the bacterial 30S ribosomal subunit.[40] There are three main mechanisms of aminoglycoside resistance mechanisms which are currently and widely accepted: aminoglycoside modifying enzymes, ribosomal mutations, and active efflux of the drug out of the bacteria.
Aminoglycoside-modifying enzymes inactivate the aminoglycoside by covalently attaching either a phosphate, nucleotide, or acetyl moiety to either the amine or the alcohol key functional group (or both groups) of the antibiotic. This changes the charge or sterically hinders the antibiotic, decreasing its ribosomal binding affinity. In S. aureus, the best-characterized aminoglycoside-modifying enzyme is aminoglycoside adenylyltransferase 4' IA (ANT(4')IA). This enzyme has been solved by x-ray crystallography.[41] The enzyme is able to attach an adenyl moiety to the 4' hydroxyl group of many aminoglycosides, including kamamycin and gentamicin.
Glycopeptide resistance is mediated by acquisition of the vanA gene. The vanA gene originates from the enterococci and codes for an enzyme that produces an alternative peptidoglycan to which vancomycin will not bind.
Today, S. aureus has become resistant to many commonly used antibiotics. In the UK, only 2% of all S. aureus isolates are sensitive to penicillin, with a similar picture in the rest of the world. The β-lactamase-resistant penicillins (methicillin, oxacillin, cloxacillin, and flucloxacillin) were developed to treat penicillin-resistant S. aureus, and are still used as first-line treatment. Methicillin was the first antibiotic in this class to be used (it was introduced in 1959), but, only two years later, the first case of MRSA was reported in England.[42]
Despite this, MRSA generally remained an uncommon finding, even in hospital settings, until the 1990s, when there was an explosion in MRSA prevalence in hospitals, where it is now endemic.[43]
MRSA infections in both the hospital and community setting are commonly treated with non-β-lactam antibiotics, such as clindamycin (a lincosamine) and co-trimoxazole (also commonly known as trimethoprim/sulfamethoxazole). Resistance to these antibiotics has also led to the use of new, broad-spectrum anti-gram-positive antibiotics, such as linezolid, because of its availability as an oral drug. First-line treatment for serious invasive infections due to MRSA is currently glycopeptide antibiotics (vancomycin and teicoplanin). There are number of problems with these antibiotics, such as the need for intravenous administration (there is no oral preparation available), toxicity, and the need to monitor drug levels regularly by blood tests. There are also concerns glycopeptide antibiotics do not penetrate very well into infected tissues (this is a particular concern with infections of the brain and meninges and in endocarditis). Glycopeptides must not be used to treat methicillin-sensitive S. aureus (MSSA), as outcomes are inferior.[44]
Because of the high level of resistance to penicillins and because of the potential for MRSA to develop resistance to vancomycin, the U.S. Centers for Disease Control and Prevention has published guidelines for the appropriate use of vancomycin. In situations where the incidence of MRSA infections is known to be high, the attending physician may choose to use a glycopeptide antibiotic until the identity of the infecting organism is known. After the infection is confirmed to be due to a methicillin-susceptible strain of S. aureus, treatment can be changed to flucloxacillin or even penicillin, as appropriate.
Vancomycin-resistant S. aureus (VRSA) is a strain of S. aureus that has become resistant to the glycopeptides. The first case of vancomycin-intermediate S. aureus (VISA) was reported in Japan in 1996;[45] but the first case of S. aureus truly resistant to glycopeptide antibiotics was only reported in 2002.[46] Three cases of VRSA infection had been reported in the United States as of 2005.[47]
Carriage of Staphylococcus aureus
The carriage of Staphylococcus aureus is an important source of nosocomial infection and community-acquired methicillin-resistant S. aureus (MRSA). Although S. aureus can be present on the skin of the host, a large proportion of its carriage is through the anterior nares of the nasal passages.[2] The ability of the nasal passages to harbour S. aureus results from a combination of a weakened or defective host immunity and the bacteria's ability to evade host innate immunity.[48]
Infection control
Spread of S. aureus (including MRSA) generally is through human-to-human contact, although recently some veterinarians have discovered the infection can be spread through pets,[49] with environmental contamination thought to play a relatively unimportant part. Emphasis on basic hand washing techniques are, therefore, effective in preventing its transmission. The use of disposable aprons and gloves by staff reduces skin-to-skin contact and, therefore, further reduces the risk of transmission. Please refer to the main article on infection control for further details.
Recently, there have been myriad reported cases of S. aureus in hospitals across America. Transmission of the pathogen is facilitated in medical settings where healthcare worker hygiene is insufficient. S. aureus is an incredibly hardy bacterium, as was shown in a study where it survived on polyester for just under three months;[50] polyester is the main material used in hospital privacy curtains.
The bacteria are transported on the hands of healthcare workers, who may pick them up from a seemingly healthy patient carrying a benign or commensal strain of S. aureus, and then pass it on to the next patient being treated. Introduction of the bacteria into the bloodstream can lead to various complications, including, but not limited to, endocarditis, meningitis, and, if it is widespread, sepsis.
Ethanol has proven to be an effective topical sanitizer against MRSA. Quaternary ammonium can be used in conjunction with ethanol to increase the duration of the sanitizing action. The prevention of nosocomial infections involves routine and terminal cleaning. Nonflammable alcohol vapor in CO
2 NAV-CO2 systems have an advantage, as they do not attack metals or plastics used in medical environments, and do not contribute to antibacterial resistance.
An important and previously unrecognized means of community-associated MRSA colonization and transmission is during sexual contact.[51]
Staff or patients who are found to carry resistant strains of S. aureus may be required to undergo "eradication therapy", which may include antiseptic washes and shampoos (such as chlorhexidine) and application of topical antibiotic ointments (such as mupirocin or neomycin) to the anterior nares of the nose.
S. aureus is killed in 1 minute at 78 °C and 10 minutes at 64 °C.[52]
The nonprotein amino acid L-homoarginine is a growth inhibitor of S. aureus as well as Candida albicans. It is assumed to be an antimetabolite of arginine.
Biological control might be a new possible way to control Staphylococcus aureus in body surfaces. Colonization of body surfaces (especially in the nose) by Staphylococcus epidermidis(inhibitory strain JK16) impairs the establishment of S. aureus.
A 2011 study[53] points to this new possible way to control S. aureus. This study was performed from observations of the nasal microbial flora of a diverse group of people. It was discovered that there are two different strains of S. epidermidis, one that inhibits biofilm formation by S. aureus, S. epidermidis strain JK16 (inhibitory type), and one that does not (non-inhibitory type) S. epidermidis strain JK11. In this study they observed that there were some patients that were not affected by Staphylococcus aureus; this was because these patients had S. aureus together with S. epidermis (inhibitory type), in their nasal microbial flora. This is due to an amensalistic relationship between these microorganisms, the inhibitory strain of S. epidermidis and Staphylococcus aureus.
These findings open the way to a biological control therapy to help in the treatment of S. aureus infections which are becoming a growing threat due to the rise of resistance to conventional antibiotic treatments.
See also
- Facultative anaerobic organism
- Gram-positive bacteria
- List of cutaneous conditions
- MRSA
- Staphylococcal infection
- Vancomycin-resistant Staphylococcus aureus
References
- ↑ Ogston A (1984). ""On Abscesses". Classics in Infectious Diseases". Rev Infect Dis 6 (1): 122–28. doi:10.1093/clinids/6.1.122. PMID 6369479.
- ↑ 2.0 2.1 2.2 2.3 Kluytmans J, van Belkum A, Verbrugh H; Van Belkum; Verbrugh (July 1997). "Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks". Clin. Microbiol. Rev. 10 (3): 505–20. PMC 172932. PMID 9227864.
- ↑ 3.0 3.1 Cole, A. M.; Tahk, S.; Oren, A.; Yoshioka, D.; Kim, Y. H.; Park, A.; Ganz, T (November 2001). "Determinants of Staphylococcus aureus nasal carriage". Clin Diagn Lab Immunol 8 (6): 1064–9. doi:10.1128/CDLI.8.6.1064-1069.2001. PMC 96227. PMID 11687441.
- ↑ Bowersox, John (27 May 1999). "Experimental Staph Vaccine Broadly Protective in Animal Studies". NIH. Archived from the original on 5 May 2007. Retrieved 28 July 2007.
- ↑ 5.0 5.1 Ryan, KJ; Ray, CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 0-8385-8529-9.
- ↑ "Staphylococcus aureus: Reproduction". palexander13.webs.com.
- ↑ PreTest, Surgery, 12th ed., p.88
- ↑ Matthews KR, Roberson J, Gillespie BE, Luther DA, Oliver SP (1997). "Identification and Differentiation of Coagulase-Negative Staphylococcus aureus by Polymerase Chain Reaction". Journal of Food Protection 60 (6): 686–8.
- ↑ Cimolai N (July 2008). "MRSA and the environment: implications for comprehensive control measures". Eur. J. Clin. Microbiol. Infect. Dis. 27 (7): 481–93. doi:10.1007/s10096-008-0471-0. PMID 18273652.
- ↑ Curran JP, Al-Salihi FL; Al-Salihi (1980). "Neonatal staphylococcal scalded skin syndrome: massive outbreak due to an unusual phage type". Pediatrics 66 (2): 285–90. PMID 6447271.
- ↑ 11.0 11.1 11.2 American Academy of Dermatology (February 2013), "Five Things Physicians and Patients Should Question", Choosing Wisely: an initiative of the ABIM Foundation (American Academy of Dermatology), retrieved 5 December 2013, which cites
- Birnie, A. J.; Bath-Hextall, F. J.; Ravenscroft, J. C.; Williams, H. C. (2008). Birnie, Andrew J, ed. "Cochrane Database of Systematic Reviews". The Cochrane database of systematic reviews (3): CD003871. doi:10.1002/14651858.CD003871.pub2. PMID 18646096.
|chapter=ignored (help)
- Birnie, A. J.; Bath-Hextall, F. J.; Ravenscroft, J. C.; Williams, H. C. (2008). Birnie, Andrew J, ed. "Cochrane Database of Systematic Reviews". The Cochrane database of systematic reviews (3): CD003871. doi:10.1002/14651858.CD003871.pub2. PMID 18646096.
- ↑ 12.0 12.1 Boost MV, O'Donoghue MM, James A; O'Donoghue; James (2008). "Prevalence of Staphylococcus aureus carriage among dogs and their owners". Epidemiol. Infect. 136 (7): 953–964. doi:10.1017/S0950268807009326. PMC 2870875. PMID 17678561.
- ↑ Hanselman BA, Kruth SA, Rousseau J, Weese JS; Kruth; Rousseau; Weese (2009). "Coagulase positive staphylococcal colonization of humans and their household pets". Can. Vet. 50 (9): 954–8. PMC 2726022. PMID 19949556.
- ↑ Burton S, Reid-Smith R, McClure JT, Weese JS; Reid-Smith; McClure; Weese (2008). "Staphylococcus aureus colonization in healthy horses in Atlantic Canada". Can. Vet. J. 49 (8): 797–799. PMC 2465786. PMID 18978975.
- ↑ "Staphylococcosis, Staphylococcal Arthritis, Bumble Foot". The Poultry Site. Retrieved 2013-10-22.
- ↑ Cenci-Goga BT, Karama M, Rossitto PV, Morgante RA, Cullor JS; Karama; Rossitto; Morgante; Cullor (September 2003). "Enterotoxin production by Staphylococcus aureus isolated from mastitic cows". Journal of food protection 66 (9): 1693–6. PMID 14503727.
- ↑ Medical Laboratory Manual For Tropical Countries vol two
- ↑ Dinges MM, Orwin PM, Schlievert PM; Orwin; Schlievert (January 2000). "Exotoxins of Staphylococcus aureus". Clin. Microbiol. Rev. 13 (1): 16–34, table of contents. doi:10.1128/cmr.13.1.16-34.2000. PMC 88931. PMID 10627489.
- ↑ Jarraud S, Peyrat MA, Lim A et al. (January 2001). "egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus". J. Immunol. 166 (1): 669–77. doi:10.4049/jimmunol.166.1.669. PMID 11123352.
- ↑ 20.0 20.1 Becker K, Friedrich AW, Lubritz G, Weilert M, Peters G, Von Eiff C; Friedrich; Lubritz; Weilert; Peters; von Eiff (April 2003). "Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens". J. Clin. Microbiol. 41 (4): 1434–9. doi:10.1128/jcm.41.4.1434-1439.2003. PMC 153929. PMID 12682126.
- ↑ Lina G, Piémont Y, Godail-Gamot F et al. (November 1999). "Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia". Clin. Infect. Dis. 29 (5): 1128–32. doi:10.1086/313461. PMID 10524952.
- ↑ Gillet Y, Issartel B, Vanhems P et al. (March 2002). "Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients". Lancet 359 (9308): 753–9. doi:10.1016/S0140-6736(02)07877-7. PMID 11888586. As PDF
- ↑ Schneewind O, Fowler A, Faull KF; Fowler; Faull (April 1995). "Structure of the cell wall anchor of surface proteins in Staphylococcus aureus". Science 268 (5207): 103–6. doi:10.1126/science.7701329. PMID 7701329.
- ↑ Patel AH, Nowlan P, Weavers ED, Foster T; Nowlan; Weavers; Foster (December 1987). "Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement". Infect. Immun. 55 (12): 3103–10. PMC 260034. PMID 3679545.
- ↑ Zhu J; Lu C; Standland M et al. (February 2008). "Single mutation on the surface of Staphylococcus aureus Sortase A can disrupt its dimerization". Biochemistry 47 (6): 1667–74. doi:10.1021/bi7014597. PMID 18193895.
- ↑ 26.0 26.1 Clauditz A, Resch A, Wieland KP, Peschel A, Götz F; Resch; Wieland; Peschel; Götz (August 2006). "Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress". Infection and immunity 74 (8): 4950–3. doi:10.1128/IAI.00204-06. PMC 1539600. PMID 16861688.
- ↑ 27.0 27.1 Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V; Essex; Buchanan; Datta; Hoffman; Bastian; Fierer; Nizet (2005). "Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity". J Exp Med 202 (2): 209–15. doi:10.1084/jem.20050846. PMC 2213009. PMID 16009720.
- ↑ Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, Nizet V, Wang AH, Oldfield E; Liu; Song; Yin; Hensler; Jeng; Nizet; Wang; Oldfield (2008). "A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence". Science 319 (5868): 391–94. doi:10.1126/science.1153018. PMC 2747771. PMID 18276850.
- ↑ Francois P and Schrenzel J (2008). "Rapid Diagnosis and Typing of Staphylococcus aureus". Staphylococcus: Molecular Genetics. Caister Academic Press. ISBN 978-1-904455-29-5.
- ↑ Mackay IM, ed. (2007). Real-Time PCR in Microbiology: From Diagnosis to Characterization. Caister Academic Press. ISBN 978-1-904455-18-9.
- ↑ 31.0 31.1 Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infection, Genetics and Evolution 2008 Dec 8;8(6): 747-763.
- ↑ Sousa MAD, Conceicao T, Simas C, Lencastre HD. Comparison of Genetic Backgrounds of Methicillin-Resistant and -Susceptible Staphylococcus aureus Isolates from Portuguese Hospitals and the Community. Journal of Clinical Microbiology 2005 Oct 5;43(10): 5150-5157.
- ↑ 33.0 33.1 33.2 Kim, J. Understanding the Evolution of Methicillin-Resistant Staphylococcus aureus. Clinical Microbiology Newsletter. 2009;31(3):17-23. doi:10.1016/j.clinmicnews.2009.01.002.
- ↑ Korzeniowski O, Sande MA; Sande (1982). "Combination antimicrobial therapy for Staphylococcus aureus endocarditis in patients addicted to parenteral drugs and in nonaddicts: a prospective study". Ann Intern Med 97 (4): 496–503. doi:10.7326/0003-4819-97-4-496. PMID 6751182.
- ↑ Bayer AS; Bolger AF; Taubert KA et al. (1998). "Diagnosis and management of infective endocarditis and its complications". Circulation 98 (25): 2936–48. doi:10.1161/01.CIR.98.25.2936. PMID 9860802.
|first10=missing|last10=in Authors list (help);|first11=missing|last11=in Authors list (help) - ↑ Cosgrove SE; Vigliani GA; Campion M et al. (2009). "Initial low‐dose gentamicin for Staphylococcus aureus bacteremia and endocarditis is nephrotoxic". Clin Infect Dis 48 (6): 713–721. doi:10.1086/597031. PMID 19207079.
- ↑ Chambers HF (2001). "The changing epidemiology of Staphylococcus aureus?". Emerg Infect Dis 7 (2): 178–82. doi:10.3201/eid0702.010204. PMC 2631711. PMID 11294701.
- ↑ Waters AE, Contente-Cuomo T, Buchhagen J et al. (May 2011). "Multidrug-Resistant Staphylococcus aureus in US Meat and Poultry". Clin. Infect. Dis. 52 (10): 1227–30. doi:10.1093/cid/cir181. PMC 3079400. PMID 21498385. [US meat and poultry is widely contaminated with drug-resistant Staph bacteria, study finds Lay summary] – ScienceDaily (15 April 2011).
- ↑ Capparelli R, Parlato M, Borriello G, Salvatore P, Iannelli D; Parlato; Borriello; Salvatore; Iannelli (August 2007). "Experimental phage therapy against Staphylococcus aureus in mice". Antimicrob. Agents Chemother. 51 (8): 2765–73. doi:10.1128/AAC.01513-06. PMC 1932491. PMID 17517843. [Phage Therapy May Control Staph Infections In Humans Including MRSA Lay summary] – ScienceDaily (22 August 2007).
- ↑ Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V; Clemons; Brodersen; Morgan-Warren; Wimberly; Ramakrishnan (September 2000). "Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics". Nature 407 (6802): 340–8. doi:10.1038/35030019. PMID 11014183.
- ↑ Sakon J, Liao HH, Kanikula AM, Benning MM, Rayment I, Holden HM; Liao; Kanikula; Benning; Rayment; Holden (November 1993). "Molecular structure of kanamycin nucleotidyltransferase determined to 3.0-A resolution". Biochemistry 32 (45): 11977–84. doi:10.1021/bi00096a006. PMID 8218273.
- ↑ Jevons MP (1961). "Celbenin-resistant staphylococci". BMJ 1 (5219): 124–5. doi:10.1136/bmj.1.5219.124-a.
- ↑ Johnson AP, Aucken HM, Cavendish S, Ganner M, Wale MC, Warner M, Livermore DM, Cookson BD; Aucken; Cavendish; Ganner; Wale; Warner; Livermore; Cookson; Uk Earss (2001). "Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS)". J Antimicrob Chemother 48 (1): 143–4. doi:10.1093/jac/48.1.143. PMID 11418528.
- ↑ Blot SI, Vandewoude KH, Hoste EA, Colardyn FA; Vandewoude; Hoste; Colardyn (2002). "Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus". Arch Intern Med 162 (19): 22293–5. doi:10.1001/archinte.162.19.2229. PMID 12390067.
- ↑ Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC; Hanaki; Ino; Yabuta; Oguri; Tenover (1997). "Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility" (PDF). J Antimicrob Chemother 40 (1): 135–6. doi:10.1093/jac/40.1.135. PMID 9249217.
- ↑ Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK; Sievert; Hageman; Boulton; Tenover; Downes; Shah; Rudrik; Pupp; Brown; Cardo; Fridkin; Vancomycin-Resistant Staphylococcus aureus Investigative Team (2003). "Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene". N Engl J Med 348 (14): 1342–7. doi:10.1056/NEJMoa025025. PMID 12672861.
- ↑ Menichetti F (2005). "Current and emerging serious Gram-positive infections". Clin Microbiol Infect 11 (Suppl 3): 22–8. doi:10.1111/j.1469-0691.2005.01138.x. PMID 15811021.
- ↑ Quinn, G. A.; Cole, A. M (September 2007). "Suppression of innate immunity by a nasal carriage strain of Staphylococcus aureus increases its colonization on nasal epithelium". Immunology 122 (1): 80–9. doi:10.1111/j.1365-2567.2007.02615.x. PMC 2265977. PMID 17472720.
- ↑ Sing A, Tuschak C, Hoermansdorfer S; Tuschak; Hörmansdorfer (March 2008). "Methicillin-Resistant Staphylococcus aureus in a Family and Its Pet Cat". N Engl J Med 358 (11): 1200–1. doi:10.1056/NEJMc0706805. PMID 18337614.
- ↑ Neely AN, Maley MP; Maley (February 2000). "Survival of enterococci and staphylococci on hospital fabrics and plastic". J. Clin. Microbiol. 38 (2): 724–6. PMC 86187. PMID 10655374.
- ↑ Cook H, Furuya E, Larson E, Vasquez G, Lowy F; Furuya; Larson; Vasquez; Lowy (2007). "Heterosexual transmission of community-associated methicillin-resistant Staphylococcus aureus". Clin Infect Dis 44 (3): 410–3. doi:10.1086/510681. PMID 17205449.
- ↑ Shafiei Y, Razavilar V, Javadi A (2011). "Thermal Death Time of Staphylococcus Aureus (PTCC=29213) and Staphylococcus Epidermidis (PTCC=1435) in Distilled Water" (PDF). Australian Journal of Basic and Applied Sciences 5 (11): 1551–4.
- ↑ Iwase, Tadayuki; Uehara, Yoshio; Shinji, Hitomi; Tajima, Akiko; Seo, Hiromi; Takada, Koji; Agata, Toshihiko; Mizunoe, Yoshimitsu (20 May 2010). "Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization" (PDF). Nature 465 (7296): 346–9. doi:10.1038/nature09074. PMID 20485435.
Further reading
Loskill, Peter; Pereira, Pedro M.; Jung, Philipp; Bischoff, Markus; Herrmann, Mathias; Pinho, Mariana G.; Jacobs, Karin (2 September 2014). "Reduction of the Peptidoglycan Crosslinking Causes a Decrease in Stiffness of the Staphylococcus aureus Cell Envelope". Biophysical Journal 107 (5): 1082–1089. doi:10.1016/j.bpj.2014.07.029. PMID 25185544. Retrieved 14 November 2014.
External links
| Wikimedia Commons has media related to Staphylococcus aureus. |
| Wikispecies has information related to: Staphylococcus aureus |
- StopMRSANow.org — Discusses how to prevent the spread of MRSA
- TheMRSA.com — Understand what the MRSA infection is all about.
- "Staphylococcus aureus". NCBI Taxonomy Browser. 1280.
- Packham, Christopher (March 16, 2015). "Successful in vivo test of breakthrough Staphylococcus aureus vaccine". Medical Press. Retrieved March 2015.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||