Stable isotope labeling by amino acids in cell culture
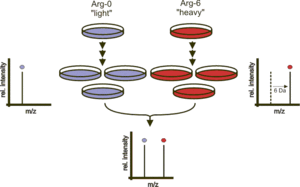
SILAC (stable isotope labeling by/with amino acids in cell culture) is a technique based on mass spectrometry that detects differences in protein abundance among samples using non-radioactive isotopic labeling.[1][2][3][4] It is a popular method for quantitative proteomics.
Procedure
Two populations of cells are cultivated in cell culture. One of the cell populations is fed with growth medium containing normal amino acids. In contrast, the second population is fed with growth medium containing amino acids labeled with stable (non-radioactive) heavy isotopes. For example, the medium can contain arginine labeled with six carbon-13 atoms (13C) instead of the normal carbon-12 (12C). When the cells are growing in this medium, they incorporate the heavy arginine into all of their proteins. Thereafter, all peptides containing a single arginine are 6 Da heavier than their normal counterparts. Alternatively, uniform labeling with 13C or 15N can be used. The trick is that the proteins from both cell populations can be combined and analyzed together by mass spectrometry. Pairs of chemically identical peptides of different stable-isotope composition can be differentiated in a mass spectrometer owing to their mass difference. The ratio of peak intensities in the mass spectrum for such peptide pairs reflects the abundance ratio for the two proteins.
Applications
A SILAC approach involving incorporation of tyrosine labeled with nine carbon-13 atoms (13C) instead of the normal carbon-12 (12C) has been utilized to study tyrosine kinase substrates in signaling pathways.[5] SILAC has emerged as a very powerful method to study cell signaling, post translation modifications such as phosphorylation,[5][6] protein–protein interaction and regulation of gene expression. In addition, SILAC has become an important method in secretomics, the global study of secreted proteins and secretory pathways.[7] It can be used to distinguish between proteins secreted by cells in culture and serum contaminants.[8] Standardized protocols of SILAC for various application have also been published.[9][10]
Pulsed SILAC
Pulsed SILAC (pSILAC) is a variation of the SILAC method where the labelled amino acids are added to the growth medium for only a short period of time. This allows monitoring differences in de novo protein production rather than raw concentration.[11]
References
- ↑ Oda Y, Huang K, Cross FR, Cowburn D, Chait BT (1999). "Accurate quantitation of protein expression and site-specific phosphorylation.". Proc Natl Acad Sci U S A 96 (12): 6591–6. doi:10.1073/pnas.96.12.6591. PMC 21959. PMID 10359756.
- ↑ Jiang H, English, AM (2002). "Quantitative Analysis of the Yeast Proteome by Incorporation of Isotopically Labeled Leucine". Journal of Proteome Research 1 (4): 345–50. doi:10.1021/pr025523f. PMID 12645890.
- ↑ Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002). "Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics". Molecular & Cellular Proteomics 1 (5): 376–86. doi:10.1074/mcp.M200025-MCP200. PMID 12118079.
- ↑ Zhu H, Pan S, Gu S, Bradbury EM, Chen X (2002). "Amino acid residue specific stable isotope labeling for quantitative proteomics". Rapid Communications in Mass Spectrometry 16 (22): 2115–23. doi:10.1002/rcm.831. PMID 12415544.
- ↑ 5.0 5.1 Ibarrola N, Molina H, Iwahori A, Pandey A (2004). "A novel proteomic approach for specific identification of tyrosine kinase substrates using 13C tyrosine". The Journal of Biological Chemistry 279 (16): 15805–13. doi:10.1074/jbc.M311714200. PMID 14739304.
- ↑ Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A (2003). "A Proteomic Approach for Quantitation of Phosphorylation Using Stable Isotope Labeling in Cell Culture". Analytical Chemistry 75 (22): 6043–49. doi:10.1021/ac034931f. PMID 14615979.
- ↑ Hathout, Yetrib (2007). "Approaches to the study of the cell secretome". Expert Review of Proteomics 4 (2): 239–48. doi:10.1586/14789450.4.2.239. PMID 17425459.
- ↑ Polacek, Martin; Bruun, Jack-Ansgar; Johansen, Oddmund; Martinez, Inigo (2010). "Differences in the secretome of cartilage explants and cultured chondrocytes unveiled by SILAC technology". Journal of Orthopaedic Research 28 (8): 1040–9. doi:10.1002/jor.21067. PMID 20108312.
- ↑ Amanchy R, Kalume DE, Pandey A (2005). "Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications". Science's STKE 2005 (16): pl2. doi:10.1126/stke.2672005pl2. PMID 15657263.
- ↑ Harsha HC, Molina H, Pandey A (2008). "Quantitative proteomics using stable isotope labeling with amino acids in cell culture". Nature Protocols 3 (3): 505–16. doi:10.1038/nprot.2008.2. PMID 18323819.
- ↑ Schwanhäusser, Björn; Manfred Gossen; Gunnar Dittmar; Matthias Selbach (January 2009). "Global analysis of cellular protein translation by pulsed SILAC". Proteomics 9 (1): 205–9. doi:10.1002/pmic.200800275. PMID 19053139. Retrieved 2009-03-17.
Further reading
- Ong SE, Kratchmarova I, Mann M (2003). "Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC)". Journal of Proteome Research 2 (2): 173–81. doi:10.1021/pr0255708. PMID 12716131.
- Ong SE, Mann M (2006). "A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC)". Nature Protocols 1 (6): 2650–60. doi:10.1038/nprot.2006.427. PMID 17406521.
- Ong SE, Mann M (2007). "Stable isotope labeling by amino acids in cell culture for quantitative proteomics". Methods in Molecular Biology 359: 37–52. doi:10.1007/978-1-59745-255-7_3. PMID 17484109.
External links
- SILAC resource Mann Lab
- SILAC resource Pandey Lab
- SILAC resource Center for Experimental Bioinformatics (CEBI)
| ||||||||||