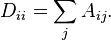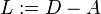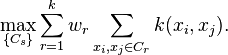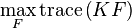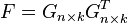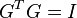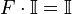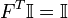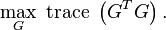Spectral clustering
In multivariate statistics and the clustering of data, spectral clustering techniques make use of the spectrum (eigenvalues) of the similarity matrix of the data to perform dimensionality reduction before clustering in fewer dimensions. The similarity matrix is provided as an input and consists of a quantitative assessment of the relative similarity of each pair of points in the dataset.
In application to image segmentation, spectral clustering is known as segmentation-based object categorization.
Algorithms
Given an enumerated set of data points, the similarity matrix may be defined as a symmetric matrix 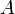 , where
, where 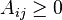 represents a measure of the similarity between data points with indexes
represents a measure of the similarity between data points with indexes 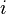 and
and 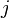 .
.
One spectral clustering technique is the normalized cuts algorithm or Shi–Malik algorithm introduced by Jianbo Shi and Jitendra Malik,[1] commonly used for image segmentation. It partitions points into two sets 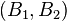 based on the eigenvector
based on the eigenvector 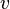 corresponding to the second-smallest eigenvalue of the
symmetric normalized Laplacian defined as
corresponding to the second-smallest eigenvalue of the
symmetric normalized Laplacian defined as
-
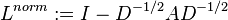 ,
,
where 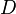 is the diagonal matrix
is the diagonal matrix
A mathematically equivalent algorithm [2] takes the eigenvector corresponding to the largest eigenvalue of the random walk normalized Laplacian matrix 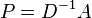 .
.
Another possibility is to use the Laplacian matrix defined as
rather than the symmetric normalized Laplacian matrix.
Partitioning may be done in various ways, such as by computing the median 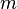 of the components of the second smallest eigenvector
of the components of the second smallest eigenvector 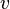 , and placing all points whose component in
, and placing all points whose component in 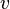 is greater than
is greater than 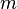 in
in 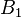 , and the rest in
, and the rest in 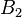 . The algorithm can be used for hierarchical clustering by repeatedly partitioning the subsets in this fashion.
. The algorithm can be used for hierarchical clustering by repeatedly partitioning the subsets in this fashion.
Alternatively to computing just one eigenvector, k eigenvectors for some k, are computed, and then another algorithm (e.g. k-means clustering) is used to cluster points by their respective k components in these eigenvectors.
The efficiency of spectral clustering may be improved if the solution to the corresponding eigenvalue problem is performed in a matrix-free fashion, i.e., without explicitly manipulating or even computing the similarity matrix, as, e.g., in the Lanczos algorithm.
For large-sized graphs, the second eigenvalue of the (normalized) graph Laplacian matrix is often ill-conditioned, leading to slow convergence of iterative eigenvalue solvers. Preconditioning is a key technology accelerating the convergence, e.g., in the matrix-free LOBPCG method. Spectral clustering has been successfully applied on large graphs by first identifying their community structure, and then clustering communities.[3]
Spectral clustering is closely related to Nonlinear dimensionality reduction, and dimension reduction techniques such as locally-linear embedding can be used to reduce errors from noise or outliers.[4]
Relationship with k-means
The kernel k-means problem is an extension of the k-means problem where the input data points are mapped non-linearly into a higher-dimensional feature space via a kernel function 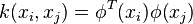 . The weighted kernel k-means problem further extends this problem by defining a weight
. The weighted kernel k-means problem further extends this problem by defining a weight 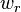 for each cluster as the reciprocal of the number of elements in the cluster,
for each cluster as the reciprocal of the number of elements in the cluster,
Suppose 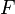 is a matrix of the normalizing coefficients for each point for each cluster
is a matrix of the normalizing coefficients for each point for each cluster 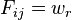 if
if 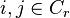 and zero otherwise. Suppose
and zero otherwise. Suppose 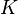 is the kernel matrix for all points. The weighted kernel k-means problem with n points and k clusters is given as,
is the kernel matrix for all points. The weighted kernel k-means problem with n points and k clusters is given as,
such that,
such that 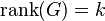 . In addition, there are identity constrains on
. In addition, there are identity constrains on 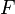 given by,
given by,
where 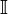 represents a vector of ones.
represents a vector of ones.
This problem can be recast as,
This problem is equivalent to the spectral clustering problem when the identity constraints on 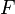 are relaxed. In particular, the weighted kernel k-means problem can be reformulated as a spectral clustering (graph partitioning) problem and vice versa. The output of the algorithms are eigenvectors which do not satisfy the identity requirements for indicator variables defined by
are relaxed. In particular, the weighted kernel k-means problem can be reformulated as a spectral clustering (graph partitioning) problem and vice versa. The output of the algorithms are eigenvectors which do not satisfy the identity requirements for indicator variables defined by 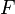 . Hence, post-processing of the eigenvectors is required for the equivalence between the problems.[5]
Transforming the spectral clustering problem into a weighted kernel k-means problem greatly reduces the computational burden.[6]
. Hence, post-processing of the eigenvectors is required for the equivalence between the problems.[5]
Transforming the spectral clustering problem into a weighted kernel k-means problem greatly reduces the computational burden.[6]
See also
References
- ↑ Jianbo Shi and Jitendra Malik, "Normalized Cuts and Image Segmentation", IEEE Transactions on PAMI, Vol. 22, No. 8, Aug 2000.
- ↑ Marina Meilă & Jianbo Shi, "Learning Segmentation by Random Walks", Neural Information Processing Systems 13 (NIPS 2000), 2001, pp. 873–879.
- ↑ Zare, Habil; P. Shooshtari, A. Gupta and R. Brinkman (2010). "Data reduction for spectral clustering to analyze high throughput flow cytometry data". BMC Bioinformatics.
- ↑ Arias-Castro, E. and Chen, G. and Lerman, G. (2011), "Spectral clustering based on local linear approximations.", Electronic Journal of Statistics 5: 1537–1587, doi:10.1214/11-ejs651
- ↑ Dhillon, I.S. and Guan, Y. and Kulis, B. (2004). "Kernel k-means: spectral clustering and normalized cuts". Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining. pp. 551–556.
- ↑ Dhillon, Inderjit; Yuqiang Guan; Brian Kulis (November 2007). "Weighted Graph Cuts without Eigenvectors: A Multilevel Approach". IEEE Transactions on Pattern Analysis and Machine Intelligence 29 (11): 1–14.