Schellman loop
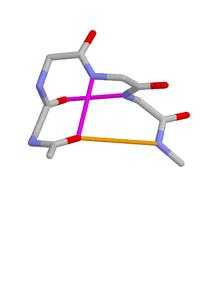
Schellman Loops (also called Schellman motifs or paperclips)[1][2][3][4][5][6][7][8][9][10] are commonly occurring structural features of proteins and polypeptides.[11] Each has six amino acid residues (labelled residues i to i+5) with two specific inter-mainchain hydrogen bonds and a characteristic main chain dihedral angle conformation. The CO group of residue i is hydrogen-bonded to the NH of residue i+5 (colored orange in Figure), and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+4 (beta turn, colored purple). Residues i+1, i+2, and i+3 have negative φ (phi) angle values and the phi value of residue i+4 is positive. Schellman loops incorporate a three amino acid residue RL nest (protein structural motif),[12] [13] in which three mainchain NH groups (from Schellman loop residues i+3 to i+5) form a concavity for hydrogen bonding to carbonyl oxygens. About 2.5% of amino acids in proteins belong to Schellman loops. Two websites are available for examining small motifs in proteins, Motivated Proteins: ;[14] or PDBeMotif: .[15]
The majority of Schellman loops (82%) occur at the C-terminus of an alpha-helix such that residues i, i+1, i+2 and i+3 are part of the helix. Over a quarter of helices (28%) have a C-terminal Schellman loop.[10]
Occasional Schellman loops occur with seven instead of six residues. In these, the CO group of residue i is hydrogen-bonded to the NH of residue i+6, and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+5. Rare “left-handed” six-residue Schellman loops occur; these have the same hydrogen bonds, but residues i+1, i+2, and i+3 have positive φ values while the φ value of residue i+4 is negative; the nest is of the LR, rather than the RL, kind.
Amino acid propensities for the six Schellman-loop residues have been described.[16] Residue i+4 is the one most-highly conserved, with 70% of amino acids being glycine and none proline.
The original Schellman criteria [1] result in the inclusion of features not now regarded as Schellman loops. A newer set of criteria is given above.
References
- ↑ 1.0 1.1 Schellman, C (1980). Protein Folding. Amsterdam: Elsevier. pp. 53–61.
- ↑ Milner-White, E J (1988-02-05). "Recurring loop motif in proteins that occurs in right-handed and left-handed forms. Its relationship with alpha-helices and beta-bulge loops". Journal of Molecular Biology 199 (3): 503–511. doi:10.1016/0022-2836(88)90621-3. ISSN 0022-2836. PMID 3351939.
- ↑ Aurora, R; Srinivasan, R; Rose, G D (1994). "Rules for alpha-helix termination by glycine". Science (New York) 264 (5162): 1126–1130. doi:10.1126/science.8178170. ISSN 0036-8075. PMID 8178170. Check date values in:
|year= / |date= mismatch(help) - ↑ Viguera, Ana Rosa; Serrano, Luis (1995-08-04). "Experimental Analysis of the Schellman Motif". Journal of Molecular Biology 251 (1): 150–160. doi:10.1006/jmbi.1995.0422. ISSN 0022-2836.
- ↑ Aurora, R; Rose, G D (January 1998). "Helix capping". Protein science: a publication of the Protein Society 7 (1): 21–38. doi:10.1002/pro.5560070103. ISSN 0961-8368. PMC 2143812. PMID 9514257.
- ↑ Kallenbach, Neville R.; Gong, Youxiang (January 1999). "C-Terminal capping motifs in model helical peptides". Bioorganic & Medicinal Chemistry 7 (1): 143–151. doi:10.1016/S0968-0896(98)00231-4. ISSN 0968-0896.
- ↑ Sukumar, Muppalla; Gierasch, Lila M (August 1997). "Local interactions in a Schellman motif dictate interhelical arrangement in a protein fragment". Folding and Design 2 (4): 211–222. doi:10.1016/S1359-0278(97)00030-8. ISSN 1359-0278.
- ↑ Datta, Saumen; Uma, Manjappara V.; Shamala, N.; Balaram, P. (1999). "Stereochemistry of Schellman motifs in peptides: crystal structure of a hexapeptide with a C-terminus 6→1 hydrogen bond". Biopolymers 50 (1): 13–22. doi:10.1002/(sici)1097-0282(199907)50:1<13::aid-bip2>3.0.co;2-k. ISSN 0006-3525.
- ↑ Sagermann, Martin; Mårtensson, Lars-Göran; Baase, Walter A.; Matthews, Brian W. (2002-03-01). "A test of proposed rules for helix capping: Implications for protein design". Protein Science 11 (3): 516–521. doi:10.1110/ps.39802. ISSN 1469-896X.
- ↑ 10.0 10.1 Leader, DP; Milner-White, EJ (2011). "The structure of the ends of alpha-helices in globular proteins". Proteins 79 (3): 1010–1019. doi:10.1002/prot.22942.
- ↑ Milner-White, E. James; Nissink, J. Willem M.; Allen, Frank H.; Duddy, William J. (2004-11-01). "Recurring main-chain anion-binding motifs in short polypeptides: nests". Acta Crystallographica Section D Biological Crystallography 60 (11): 1935–1942. doi:10.1107/S0907444904021390. ISSN 0907-4449.
- ↑ Datta, S; Uma, MV (1999). "Stereochemistry of Schellman motifs in peptides. Crystal structure of a hexapeptide with a C-terminal 6->1 hydrogen bond". Biopolymers 50 (1): 13–22. doi:10.1002/(sici)1097-0282(199907)50:1<13::aid-bip2>3.0.co;2-k.
- ↑ Watson, James D; Milner-White, E James (2002-01-11). "A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi,psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions". Journal of Molecular Biology 315 (2): 171–182. doi:10.1006/jmbi.2001.5227. ISSN 0022-2836. PMID 11779237.
- ↑ Leader, DP; Milner-White (2009). "Motivated Proteins: A web application for studying small three-dimensional protein motifs". BMC Bioinformatics 10 (1): 60. doi:10.1186/1471-2105-10-60. PMC 2651126. PMID 19210785.
- ↑ Golovin, A; Henrick (2008). "MSDmotif: exploring protein sites and motifs". BMC Bioinformatics 9 (1): 312. doi:10.1186/1471-2105-9-312. PMC 2491636. PMID 18637174.
- ↑ Newell, Nicholas E (2011-12-15). "Cascade detection for the extraction of localized sequence features; specificity results for HIV-1 protease and structure-function results for the Schellman loop". Bioinformatics (Oxford, England) 27 (24): 3415–3422. doi:10.1093/bioinformatics/btr594. ISSN 1367-4811. PMID 22039211.