Ribokinase
| ribokinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.7.1.15 | ||||||||
| CAS number | 9026-84-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
In enzymology, a ribokinase (EC 2.7.1.15) is an enzyme that catalyzes the chemical reaction
- ATP + D-ribose
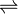 ADP + D-ribose 5-phosphate
ADP + D-ribose 5-phosphate
Thus, the two substrates of this enzyme are ATP and D-ribose, whereas its two products are ADP and D-ribose 5-phosphate.
This enzyme belongs to the family of transferases, specifically those transferring phosphorus-containing groups (phosphotransferases) with an alcohol group as acceptor. The systematic name of this enzyme class is ATP:D-ribose 5-phosphotransferase. Other names in common use include deoxyribokinase, ribokinase (phosphorylating), and D-ribokinase. This enzyme participates in pentose phosphate pathway.
Structural studies
As of late 2007, 7 structures have been solved for this class of enzymes, with PDB accession codes 1GQT, 1RK2, 1RKA, 1RKD, 1RKS, 1VM7, and 2FV7.
References
- Agranoff BW and Brady RO (1956). "Purification and properties of calf liver ribokinase". J. Biol. Chem. 219: 221–229.
- GINSBURG A (1959). "A deoxyribokinase from Lactobacillus plantarum". J. Biol. Chem. 234 (3): 481–7. PMID 13641245.