Rhizobium leguminosarum exopolysaccharide glucosyl ketal-pyruvate-transferase
| Rhizobium leguminosarum exopolysaccharide glucosyl ketal-pyruvate-transferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.5.1.98 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Rhizobium leguminosarum exopolysaccharide glucosyl ketal-pyruvate-transferase (EC 2.5.1.98, PssM) is an enzyme with system name phosphoenolpyruvate:(D-GlcA-beta-(1->4)-2-O-Ac-D-GlcA-beta-(1->4)-D-Glc-beta-(1->4)-(3-O-CH3-CH2CH(OH)C(O)-D-Gal-beta-(1->4)-D-Glc-beta-(1->4)-D-Glc-beta-(1->4)-D-Glc-beta-(1->6))-2(or3)-O-Ac-D-Glc-alpha-(1->6))n 4,6-O-(1-carboxyethan-1,1-diyl)transferase .[1] This enzyme catalyses the following chemical reaction
- phosphoenolpyruvate + [D-GlcA-beta-(1->4)-2-O-Ac-D-GlcA-beta-(1->4)-D-Glc-beta-(1->4)-[3-O-CH3-CH2CH(OH)C(O)-D-Gal-beta-(1->4)-D-Glc-beta-(1->4)-D-Glc-beta-(1->4)-D-Glc-beta-(1->6)]-2(or3)-O-Ac-D-Glc-alpha-(1->6)]n
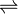 [D-GlcA-beta-(1->4)-2-O-Ac-D-GlcA-beta-(1->4)-D-Glc-beta-(1->4)-[3-O-CH3-CH2CH(OH)C(O)-D-Gal-beta-(1->3)-4,6-CH3(COO-)C-D-Glc-beta-(1->4)-D-Glc-beta-(1->4)-D-Glc-beta-(1->6)]-2(or3)-O-Ac-D-Glc-alpha-(1->6)]n + phosphate
[D-GlcA-beta-(1->4)-2-O-Ac-D-GlcA-beta-(1->4)-D-Glc-beta-(1->4)-[3-O-CH3-CH2CH(OH)C(O)-D-Gal-beta-(1->3)-4,6-CH3(COO-)C-D-Glc-beta-(1->4)-D-Glc-beta-(1->4)-D-Glc-beta-(1->6)]-2(or3)-O-Ac-D-Glc-alpha-(1->6)]n + phosphate
The enzyme is responsible for pyruvylation of subterminal glucose in the acidic octasaccharide repeating unit of the exopolysaccharide of Rhizobium leguminosarum.
References
- ↑ Ivashina, T.V., Fedorova, E.E., Ashina, N.P., Kalinchuk, N.A., Druzhinina, T.N., Shashkov, A.S., Shibaev, V.N. and Ksenzenko, V.N. (2010). "Mutation in the pssM gene encoding ketal pyruvate transferase leads to disruption of Rhizobium leguminosarum bv. viciae—Pisum sativum symbiosis". J. Appl. Microbiol. 109 (2): 731–742. doi:10.1111/j.1365-2672.2010.04702.x. PMID 20233262.
External links
- Rhizobium leguminosarum exopolysaccharide glucosyl ketal-pyruvate-transferase at the US National Library of Medicine Medical Subject Headings (MeSH)