Photosystem II

Photosystem II (or water-plastoquinone oxidoreductase) is the first protein complex in the light-dependent reactions of oxygenic photosynthesis. It is located in the thylakoid membrane of plants, algae, and cyanobacteria. Within the photosystem, enzymes capture photons of light to energize electrons that are then transferred through a variety of coenzymes and cofactors to reduce plastoquinone to plastoquinol. The energized electrons are replaced by oxidizing water to form hydrogen ions and molecular oxygen.
By replenishing lost electrons with electrons from the splitting of water, photosystem II provides the electrons for all of photosynthesis to occur. The hydrogen ions (protons) generated by the oxidation of water help to create a proton gradient that is used by ATP synthase to generate ATP. The energized electrons transferred to plastoquinone are ultimately used to reduce NADP+
to NADPH or are used in cyclic photophosphorylation.[1]
Structure
The core of PSII consists of a pseudo-symmetric heterodimer of two homologous proteins D1 and D2.[2] Unlike the reaction centers of all other photosystems which have a special pair of closely spaced chlorophyll molecules, the pigment that undergoes the initial photoinduced charge separation in PSII is a chlorophyll monomer. Because the positive charge is not shared across two molecules, the ionised pigment is highly oxidizing and can take part in the splitting of water.[2]
Photosystem II (of cyanobacteria and green plants) is composed of around 20 subunits (depending on the organism) as well as other accessory, light-harvesting proteins. Each photosystem II contains at least 99 cofactors: 35 chlorophyll a, 12 beta-carotene, two pheophytin, two plastoquinone, two heme, one bicarbonate, 20 lipid, the Mn
4CaO
5 cluster (including two chloride ions), and one non heme Fe2+
and two putative Ca2+
ion per monomer.[3] There are different crystal structures of photosystem II. The PDB accession codes for this protein are 3ARC, 3BZ1, 3BZ2 (3BZ1 and 3BZ2 are monomeric structures of the Photosystem II dimer) [3] 2AXT, 1S5L, 1W5C, 1ILX, 1FE1, 1IZL.
| Photosystem II Light Harvesting complex protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | PSII | ||||||||
| Pfam | PF00421 | ||||||||
| InterPro | IPR000932 | ||||||||
| TCDB | 3.E.2 | ||||||||
| OPM superfamily | 2 | ||||||||
| OPM protein | 3arc | ||||||||
| |||||||||
| Protein Subunits (only with known function) | |
|---|---|
| Subunit | Function |
| D1 | Reaction center Protein, binds Chlorophyll P680, pheophytin, beta-carotene,quinone and manganese center |
| D2 | Reaction center Protein |
| CP43 | Binds manganese center |
| CP47 | |
| PsbO | Manganese Stabilizing Protein |
| Coenzymes/Cofactors | |
| Molecule | Function |
| Chlorophyll | Absorbs light energy and converts it to chemical energy |
| Beta-Carotene | quench excess photoexcitation energy |
| Heme b559 | also Protoporphyrin IX containing iron |
| Pheophytin | Primary electron acceptor |
| Plastoquinone | Mobile intra-thylakoid membrane electron carrier |
| Manganese center | also known as the oxygen evolving center, or OEC |
Oxygen-evolving complex (OEC)
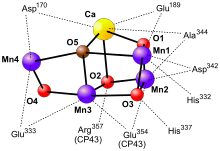
The oxygen-evolving complex is the site of water oxidation. It is a metallo-oxo cluster comprising four manganese ions (in oxidation states ranging from +3 to +4) and one divalent calcium ion. When it oxidizes water, producing oxygen gas and protons, it sequentially delivers the four electrons from water to a tyrosine (D1-Y161) sidechain and then to P680 itself. The structure of the oxygen-evolving complex is still contentious. The structures obtained by X-ray crystallography are particularly controversial, since there is evidence that the manganese atoms are reduced by the high-intensity X-rays used, altering the observed OEC structure. However, crystallography in combination with a variety of other (less damaging) spectroscopic methods such as EXAFS and electron paramagnetic resonance have given a fairly clear idea of the structure of the cluster. One possibility is the cubane-like structure.[4] In 2011 the OEC of PSII was resolved to a level of 1.9 angstroms revealing five oxygen atoms serving as oxo bridges linking the five metal atoms and four water molecules bound to the Mn4CaO5 cluster; more than 1,300 water molecules were found in each photosystem II monomer, some forming extensive hydrogen-bonding networks that may serve as channels for protons, water or oxygen molecules.[5]
Water splitting

Photosynthetic water splitting (or oxygen evolution) is one of the most important reactions on the planet, since it is the source of nearly all the atmosphere's oxygen. Moreover, artificial photosynthetic water-splitting may contribute to the effective use of sunlight as an alternative energy-source.
The mechanism of water oxidation is still not fully elucidated, but we know many details about this process. The oxidation of water to molecular oxygen requires extraction of four electrons and four protons from two molecules of water. The experimental evidence that oxygen is released through cyclic reaction of oxygen evolving complex (OEC) within one PSII was provided by Pierre Joliot et al.[6] They have shown that, if dark-adapted photosynthetic material (higher plants, algae, and cyanobacteria) is exposed to a series of single turnover flashes, oxygen evolution is detected with typical period-four damped oscillation with maxima on the third and the seventh flash and with minima on the first and the fifth flash (for review see [7]). Based on this experiment, Bessel Kok and co-workers [8] introduced a cycle of five flash-induced transitions of the so-called S-states, describing the four redox states of OEC: When four oxidizing equivalents have been stored (at the S4-state), OEC returns to its basic and in the dark stable S0-state. Finally, the intermediate S-states [9] were proposed by Jablonsky and Lazar as a regulatory mechanism and link between S-states and tyrosine Z.
See also
- Oxygen evolution
- P680
- Photosynthesis
- Photosystem
- Photosystem I
- Photosystem II light-harvesting protein
- Reaction Centre
- Photoinhibition
References
- ↑ Loll B, Kern J, Saenger W, Zouni A, Biesiadka J (December 2005). "Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II". Nature 438 (7070): 1040–4. doi:10.1038/nature04224. PMID 16355230.
- ↑ 2.0 2.1 Rutherford AW, Faller P (2003-01-29). "Photosystem II: evolutionary perspectives". Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 358 (1429): 245–253. doi:10.1098/rstb.2002.1186. PMC 1693113. PMID 12594932.
- ↑ 3.0 3.1 Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W (March 2009). "Cyanobacterial photosystem II at 2.9 Å resolution and the role of quinones, lipids, channels and chloride". Nat. Struct. Mol. Biol. 16 (3): 334–42. doi:10.1038/nsmb.1559. PMID 19219048.
- ↑ Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (March 2004). "Architecture of the photosynthetic oxygen-evolving center". Science 303 (5665): 1831–8. doi:10.1126/science.1093087. PMID 14764885.
- ↑ Umena Y, Kawakami K, Shen JR, Kamiya N (2011). "Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å". Nature 473 (7345): 55–60. doi:10.1038/nature09913. PMID 21499260.
- ↑ Joliot P., Barbieri G., Chabaud R. (1969). "Un nouveau modele des centres photochimiques du systeme II". Photochemistry and Photobiology 10 (5): 309–329. doi:10.1111/j.1751-1097.1969.tb05696.x.
- ↑ Joliot P (2003). "Period-four oscillations of the flash-induced oxygen formation in photosynthesis". Photosyn. Res. 76 (1-3): 65–72. doi:10.1023/A:1024946610564. PMID 16228566.
- ↑ Kok B, Forbush B, McGloin M (June 1970). "Cooperation of charges in photosynthetic O2 evolution-I. A linear four step mechanism". Photochem. Photobiol. 11 (6): 457–75. doi:10.1111/j.1751-1097.1970.tb06017.x. PMID 5456273.
- ↑ Jablonsky J, Lazar D (April 2008). "Evidence for intermediate S-states as initial phase in the process of oxygen-evolving complex oxidation". Biophys. J. 94 (7): 2725–36. doi:10.1529/biophysj.107.122861. PMC 2267143. PMID 18178650.