Paleopolyploidy

Paleopolyploidy is the result of genome duplications which occurred at least several million years ago (MYA). Such an event could either double the genome of a single species (autopolyploidy) or combine those of two species (allopolyploidy). Because of functional redundancy, genes are rapidly silenced or lost from the duplicated genomes. Most paleopolyploids, through evolutionary time, have lost their polyploid status through a process called diploidization, and are currently considered diploids (e.g. baker's yeast,[1] Arabidopsis thaliana,[2] and perhaps humans[3]).
Paleopolyploidy is extensively studied in plant lineages. It has been found that almost all flowering plants have undergone at least one round of genome duplication at some point during their evolutionary history. Ancient genome duplications are also found in the early ancestor of vertebrates (which includes the human lineage) and another near the origin of the bony fishes. Evidence suggests that baker's yeast (Saccharomyces cerevisiae), which has a compact genome, experienced polyploidization during its evolutionary history.
The term mesopolyploid is sometimes used for species that have undergone whole genome multiplication events (whole genome duplication, whole genome triplification, etc.) in more recent history, such as within the last 17 million years.[4]
Eukaryotes

Ancient genome duplications are widespread throughout eukaryotic lineages, particularly in plants. Studies suggest that the common ancestor of Poaceae, the grass family which includes important crop species such as maize, rice, wheat, and sugar cane, shared a whole genome duplication about 70 million years ago.[5] In more ancient monocot lineages one or likely multiple rounds of additional whole genome duplications had occurred, which were however not shared with the ancestral eudicots.[6] Further independent more recent whole genome duplications have occurred in the lineages leading to maize, sugar cane and wheat, but not rice, sorghum or foxtail millet.
A polyploidy event 160 million years ago is theorized to have created the ancestral line that led to all modern flowering plants.[7] That paleopolyploidy event was studied by sequencing the genome of an ancient flowering plant, Amborella trichopoda.[8]
The core eudicots also shared a common whole genome triplication (paleo-hexaploidy), which was estimated to have occurred after monocot-eudicot divergence but before the divergence of rosids and asterids.[9][10][11] Many eudicot species have experienced additional whole genome duplications or triplications. For example, the model plant Arabidopsis thaliana, the first plant to have its entire genome sequenced, has experienced at least two additional rounds of whole genome duplication since the duplication shared by the core eudicots.[2] The most recent event took place before the divergence of the Arabidopsis and Brassica lineages, about 20 million years ago to 45 million years ago. Other examples include the sequenced eudicot genomes of apple, soybean, tomato, cotton, etc.
Compared with plants, paleopolyploidy is much rarer in the animal kingdom. It has been identified mainly in amphibians and bony fishes. Although some studies suggested one or more common genome duplications are shared by all vertebrates (including humans), the evidence is not as strong as in the other cases because the duplications, if they exist, happened so long ago, and the matter is still under debate. The idea that vertebrates share a common whole genome duplication is known as the 2R Hypothesis. Many researchers are interested in the reason why animal lineages, particularly mammals, have had so many fewer whole genome duplications than plant lineages.
A well-supported paleopolyploidy has been found in baker's yeast (Saccharomyces cerevisiae), despite its small, compact genome (~13Mbp), after the divergence from the common yeast Kluyveromyces waltii.[12] Through genome streamlining, yeast has lost 90% of the duplicated genome over evolutionary time and is now recognized as a diploid organism.
Detection method
Duplicated genes can be identified through sequence homology on the DNA or protein level. Paleopolyploidy can be identified as massive gene duplication at one time using a molecular clock. To distinguish between whole-genome duplication and a collection of (more common) single gene duplication events, the following rules are often applied:
- Duplicated genes are located in large duplicated blocks. Single gene duplication is a random process and tends to make duplicated genes scattered throughout the genome.
- Duplicated blocks are non-overlapping because they were created simultaneously. Segmental duplication within the genome can fulfill the first rule; but multiple independent segmental duplications could overlap each other.
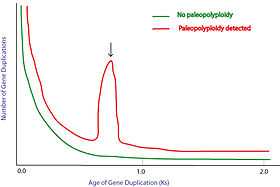
In theory, the two duplicated genes should have the same "age"; that is, the divergence of the sequence should be equal between the two genes duplicated by paleopolyploidy (homeologs). Synonymous substitution rate, Ks, is often used as a molecular clock to determine the time of gene duplication. Thus, paleopolyploidy is identified as a "peak" on the duplicate number vs. Ks graph (shown on the right).
Duplication events that occurred a long time ago in the history of various evolutionary lineages can be difficult to detect because of subsequent diploidization (such that a polyploid starts to behave cytogenetically as a diploid over time) as mutations and gene translations gradually make one copy of each chromosome unlike its counterpart. This usually results in a low confidence for identifying a very ancient paleopolyploidy.
Evolutionary importance
Paleopolyploidization events lead to massive cellular changes, including doubling of the genetic material, changes in gene expression and increased cell size. Gene loss during diploidization is not completely random, but heavily selected. Genes from large gene families are duplicated. On the other hand, individual genes are not duplicated. Overall, paleopolyploidy can have both short-term and long-term evolutionary effects on an organism's fitness in the natural environment.
- Genome diversity
- Genome doubling provided the organism with redundant alleles that can evolve freely with little selection pressure. The duplicated genes can undergo neofunctionalization or subfunctionalization which could help the organism adapt to the new environment or survive different stress conditions.
- Heterosis
- Polyploids often have larger cells and even larger organs. Many important crops, including wheat, maize and cotton, are paleopolyploids which were selected for domestication by ancient peoples.
- Speciation
- It has been suggested that many polyploidization events created new species, via a gain of adaptive traits, or by sexual incompatibility with their diploid counterparts. An example would be the recent speciation of allopolyploid Spartina — S. anglica; the polyploid plant is so successful that it is listed as an invasive species in many regions.
Vertebrates as paleopolyploid
The hypothesis of vertebrate paleopolyploidy originated as early as the 1970s, proposed by the biologist Susumu Ohno. He reasoned that the vertebrate genome could not achieve its complexity without large scale whole-genome duplications. The "two rounds of genome duplication" hypothesis (2R hypothesis) came about, and gained in popularity, especially among developmental biologists.
Some researchers have questioned the 2R hypothesis because it predicts that vertebrate genomes should have a 4:1 gene ratio compared with invertebrate genomes, and this is not supported by findings from the 48 vertebrate genome projects available in mid-2011. For example, the human genome consists of ~21,000 protein coding genes according to June, 2011 counts at UCSC and Ensembl genome analysis centers while an average invertebrate genome size is about 15,000 genes. However, the recent completion of the amphioxus genome sequence provides support for the hypothesis of two rounds of whole genome duplication, followed by loss of duplicate copies of most genes.[13] Additional arguments against 2R were based on the lack of the (AB)(CD) tree topology amongst four members of a gene family in vertebrates. However, if the two genome duplications occurred close together, we would not expect to find this topology.[14]
See also
- Gene duplication
- Genomics
- Karyotype
- Ploidy
- Polyploidy
- Speciation
References
- ↑ Kellis, M.; Birren, B. W.; Lander, E. S. (2004). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature 428 (6983): 617–624. doi:10.1038/nature02424.
- ↑ 2.0 2.1 Bowers, J. E.; Chapman, B. A.; Rong, J.; Paterson, A. H. (2003). "Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events". Nature 422 (6930): 433–438. doi:10.1038/nature01521. PMID 12660784.
- ↑ Smith, J. J., Kuraku, S., Holt, C., Sauka-Spengler, T., Jiang, N., Campbell, M. S., . . . Li, W. (2013). Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. doi:10.1038/ng.2568
- ↑ Xiaowu Wang et al. (October 2011). "The genome of the mesopolyploid crop species Brassica rapa". Nature genetics 43 (10): 1035–1039. doi:10.1038/ng.919. PMID 21873998.
- ↑ Paterson, A. H.; Bowers, J. E.; Chapman, B. A. (2004). "Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics". Proceedings of the National Academy of Sciences 101 (26): 9903. doi:10.1073/pnas.0307901101.
- ↑ Tang, H.; Bowers, J. E.; Wang, X.; Paterson, A. H. (2009). "Angiosperm genome comparisons reveal early polyploidy in the monocot lineage". Proceedings of the National Academy of Sciences 107: 472. doi:10.1073/pnas.0908007107.
- ↑ Ewen Callaway (December 2013). "Shrub genome reveals secrets of flower power". Nature. doi:10.1038/nature.2013.14426.
- ↑ Keith Adams (December 2013). "Genomic Clues to the Ancestral Flowering Plant". Science 342 (6165): 1456–1457. doi:10.1126/science.1248709.
- ↑ Tang, H.; Wang, X.; Bowers, J. E.; Ming, R.; Alam, M.; Paterson, A. H. (2008). "Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps". Genome Res 18 (12): 1944–1954. doi:10.1101/gr.080978.108.
- ↑ Jaillon O. et al. (September 2007). "The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla". Nature 449 (7161): 463–7. Bibcode:2007Natur.449..463J. doi:10.1038/nature06148. PMID 17721507.
- ↑ Tang, H.; Bowers, J. E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A. H. (2008). "Synteny and collinearity in plant genomes". Science 320 (5875): 486–488. doi:10.1126/science.1153917.
- ↑ Wong, S.; Butler, G.; Wolfe, K. H. (2002). "Gene order evolution and paleopolyploidy in hemiascomycete yeasts". Proceedings of the National Academy of Sciences 99 (14): 9272–9277. doi:10.1073/pnas.142101099. PMC 123130. PMID 12093907.
- ↑ Putnam NH, Butts T, Ferrier DE et al. (June 2008). "The amphioxus genome and the evolution of the chordate karyotype". Nature 453 (7198): 1064–71. Bibcode:2008Natur.453.1064P. doi:10.1038/nature06967. PMID 18563158.
- ↑ Furlong RF, Holland PW (April 2002). "Were vertebrates octoploid?". Philos. Trans. R. Soc. Lond., B, Biol. Sci. 357 (1420): 531–44. doi:10.1098/rstb.2001.1035. PMC 1692965. PMID 12028790.
Further reading
- Adams KL, Wendel JF (April 2005). "Polyploidy and genome evolution in plants". Curr. Opin. Plant Biol. 8 (2): 135–41. doi:10.1016/j.pbi.2005.01.001. PMID 15752992.
- Cui L, Wall PK, Leebens-Mack JH et al. (June 2006). "Widespread genome duplications throughout the history of flowering plants". Genome Res. 16 (6): 738–49. doi:10.1101/gr.4825606. PMC 1479859. PMID 16702410.
- Wolfe KH (May 2001). "Yesterday's polyploids and the mystery of diploidization". Nat. Rev. Genet. 2 (5): 333–41. doi:10.1038/35072009. PMID 11331899.
- Blanc G, Wolfe KH (July 2004). "Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes". Plant Cell 16 (7): 1667–78. doi:10.1105/tpc.021345. PMC 514152. PMID 15208399.
- Blanc G, Wolfe KH (July 2004). "Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution". Plant Cell 16 (7): 1679–91. doi:10.1105/tpc.021410. PMC 514153. PMID 15208398.
- Comai L (November 2005). "The advantages and disadvantages of being polyploid". Nat. Rev. Genet. 6 (11): 836–46. doi:10.1038/nrg1711. PMID 16304599.
- Otto SP, Whitton J (2000). "Polyploid incidence and evolution". Annu. Rev. Genet. 34 (1): 401–437. doi:10.1146/annurev.genet.34.1.401. PMID 11092833.
- Makalowski W (May 2001). "Are we polyploids? A brief history of one hypothesis". Genome Res. 11 (5): 667–70. doi:10.1101/gr.188801. PMID 11337465.
- Kellis M, Birren BW, Lander ES (April 2004). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature 428 (6983): 617–24. Bibcode:2004Natur.428..617K. doi:10.1038/nature02424. PMID 15004568.
| ||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||
| ||||||||||