PTPN1
Tyrosine-protein phosphatase non-receptor type 1 also known as protein-tyrosine phosphatase 1B (PTP1B) is an enzyme that is the founding member of the protein tyrosine phosphatase (PTP) family. In humans it is encoded by the PTPN1 gene.[1] PTP1B is a negative regulator of the insulin signaling pathway and is considered a promising potential therapeutic target, in particular for treatment of type 2 diabetes.[2] It has also been implicated in the development of breast cancer and has been explored as a potential therapeutic target in that avenue as well.[3][4][5]
Structure and function
PTP1B was first isolated from a human placental protein extract,[6][7] but it is expressed in many tissues.[8] PTP1B is localized to the cytoplasmic face of the endoplasmic reticulum.[9] PTP1B can dephosphorylate the phosphotyrosine residues of the activated insulin receptor kinase.[7][10][11] In mice, genetic ablation of PTPN1 results in enhanced insulin sensitivity.[12][13] Several other tyrosine kinases, including epidermal growth factor receptor,[14] insulin-like growth factor 1 receptor,[15] colony stimulating factor 1 receptor,[16] c-Src,[17] Janus kinase 2,[18] TYK2,[18] and focal adhesion kinase[19] as well as other tyrosine-phosphorylated proteins, including BCAR1,[20] DOK1,[21] beta-catenin[22] and cortactin[23] have also been described as PTP1B substrates.
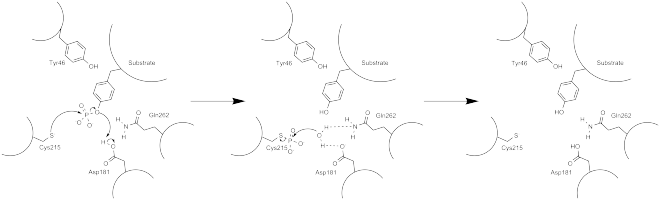
The first crystal structure of the PTP1B catalytic domain revealed that the catalytic site exists within a deep cleft of the protein formed by three loops including the WPD loop with the Asp181 residue, a pTyr loop with the Tyr46 residue and a Q loop with the Gln262 residue.[24][25] The pTyr loop and Tyr46 residue are located on the surface of the protein, and thus help to determine the depth a substrate can obtain within the cleft. This acts as a means of driving selectivity, as substrates containing smaller phosphoresidues cannot reach the site of catalytic activity at the base of the cleft.[24] Upon substrate binding, PTP1B undergoes a structural modification in which the WPD loop closes around the substrate, introducing stabilizing pi stacking interactions between the aromatic rings of the phosphotyrosine (pTyr) substrate residue and the Phe182 residue on the WPD loop.[25]
Mechanism
The phosphatase activity of PTP1B occurs via a two-step mechanism.[24] The dephosphorylation of the pTyr substrate occurs in the first step, while the enzyme intermediates are broken down during the second step. During the first step, there is a nucleophilic attack at the phosphocenter by the reduced Cys215 residue, followed by subsequent protonation by Asp181 to yield the neutral tyrosine phenol. The active enzyme is regenerated after the thiophosphate intermediate is hydrolyzed, which is facilitated by the hydrogen bonding interactions of Gln262 and Asp181 that help to position in the water molecule at the desired site of nucleophillic attack.
Regulation
The Cys215 residue is essential for the enzymatic activity of PTP1B and similar cysteine residues are required for the activity of other members of the Class I PTP family.[26] The thiolate anion form is needed for nucleophilic activity but it is susceptible to oxidation by reactive oxygen species (ROS) in the cell which would render the enzyme non-functional. This cysteine residue has been shown to oxidize under increased cellular concentrations of hydrogen peroxide (H2O2), produced in response to EGF and insulin signaling.[27][28][29] The thiolate is oxidized to a sulfenic acid, which is converted to a sulfenyl amide after reacting the with the adjacent Ser216 residue.[30] This modification of the Cys215 residue prevents further oxidation of the residue which would be irreversible, and also induces a structural change in the cleft of the active site such that substrates may not bind.[30][31] This oxidation can be reversed through reduction by glutathione and acts as a means of regulating PTP1B activity.[31] Phosphprylation of the Ser50 residue has also been shown as a point of allosteric regulation of PTP1B, in which the phosphorylated state of the enzyme is inactive.[32]
Interactions
PTPN1 has been shown to interact with BCAR1,[20] epidermal growth factor receptor,[33][34] Grb2[20][35] and IRS1.[32][35]
Clinical Significance
PTP1B has clinical implications in the treatment of type 2 diabetes as well as cancer. Gene knockout studies conducted in murine models has provided substantial evidence for the role PTP1B plays in the regulation of insulin signalling and the development of obesity.[12][13] PTPN1 knockout mice kept on high fat diets showed a resistance to obesity and an increased degree of insulin sensitivity as compared to their wild-type counterparts.[12][13] As such, the design and development of PTP1B inhibitors is a growing field of research for the treatment of type 2 diabetes and obesity.[36]
Although PTP1B is generally studied as a regulator of metabolism, some research suggest it may have a role in tumor development, though whether it is oncogenic or tumor suppressive is unclear, as there is data in support of both arguments. The high ROS concentrations within cancer cells provide an environment for potential constitutive inactivation of PTP1B and it has been shown in two human cancer cell lines HepG2 and A431, that up to 40% of the Cys215 residues in PTP1B can be selectively irreversibly oxidized under these cellular conditions resulting in non-functional PTP1B.[37] In addition, PTPN1 genetic ablation in p53 deficient mice resulted in an increased incidence of lymphomas and a decrease in overall survival rates.[38] In contrast, the PTPN1 gene has been shown to be overexpressed in conjunction with HER2 in breast cancer cases.[4] Murine models of HER2 overexpression in conjunction with PTPN1 knockout resulted in delayed tumor growth and with fewer observed metastases to the lung suggesting that PTPN1 may have an oncogenic role in breast cancer.[4][5]
See also
References
- ↑ Brown-Shimer S, Johnson KA, Lawrence JB, Johnson C, Bruskin A, Green NR et al. (Aug 1990). "Molecular cloning and chromosome mapping of the human gene encoding protein phosphotyrosyl phosphatase 1B". Proc Natl Acad Sci USA 87 (13): 5148–52. doi:10.1073/pnas.87.13.5148. PMC 54279. PMID 2164224.
- ↑ Combs AP (March 2010). "Recent advances in the discovery of competitive protein tyrosine phosphatase 1B inhibitors for the treatment of diabetes, obesity, and cancer". J. Med. Chem. 53 (6): 2333–44. doi:10.1021/jm901090b. PMID 20000419.
- ↑ Lessard L, Stuible M, Tremblay ML (2010). "The two faces of PTP1B in cancer". Biochim. Biophys. Acta 1804 (3): 613–9. doi:10.1016/j.bbapap.2009.09.018. PMID 19782770.
- ↑ 4.0 4.1 4.2 Bentires-Alj M, Neel BG (2007). "Protein-tyrosine phosphatase 1B is required for HER2/Neu-induced breast cancer". Cancer Res. 67 (6): 2420–4. doi:10.1158/0008-5472.CAN-06-4610. PMID 17347513.
- ↑ 5.0 5.1 Julien SG, Dubé N, Read M, Penney J, Paquet M, Han Y et al. (2007). "Protein tyrosine phosphatase 1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis and protects from lung metastasis". Nat. Genet. 39 (3): 338–46. doi:10.1038/ng1963. PMID 17259984.
- ↑ Tonks NK, Diltz CD, Fischer EH (May 1988). "Purification of the major protein-tyrosine-phosphatases of human placenta" (PDF). J. Biol. Chem. 263 (14): 6722–30. PMID 2834386.
- ↑ 7.0 7.1 Tonks NK, Diltz CD, Fischer EH (May 1988). "Characterization of the major protein-tyrosine-phosphatases of human placenta" (PDF). J. Biol. Chem. 263 (14): 6731–7. PMID 2834387.
- ↑ Chernoff J, Schievella AR, Jost CA, Erikson RL, Neel BG (April 1990). "Cloning of a cDNA for a major human protein-tyrosine-phosphatase". Proc. Natl. Acad. Sci. U.S.A. 87 (7): 2735–9. doi:10.1073/pnas.87.7.2735. PMC 53765. PMID 2157211.
- ↑ Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG (February 1992). "The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence". Cell 68 (3): 545–60. doi:10.1016/0092-8674(92)90190-N. PMID 1739967.
- ↑ Cicirelli MF, Tonks NK, Diltz CD, Weiel JE, Fischer EH, Krebs EG (July 1990). "Microinjection of a protein-tyrosine-phosphatase inhibits insulin action in Xenopus oocytes". Proc. Natl. Acad. Sci. U.S.A. 87 (14): 5514–8. doi:10.1073/pnas.87.14.5514. PMC 54355. PMID 2164686.
- ↑ Seely BL, Staubs PA, Reichart DR, Berhanu P, Milarski KL, Saltiel AR et al. (October 1996). "Protein tyrosine phosphatase 1B interacts with the activated insulin receptor". Diabetes 45 (10): 1379–85. doi:10.2337/diabetes.45.10.1379. PMID 8826975.
- ↑ 12.0 12.1 12.2 Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL et al. (March 1999). "Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene". Science 283 (5407): 1544–8. doi:10.1126/science.283.5407.1544. PMID 10066179.
- ↑ 13.0 13.1 13.2 Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM et al. (August 2000). "Increased Energy Expenditure, Decreased Adiposity, and Tissue-Specific Insulin Sensitivity in Protein-Tyrosine Phosphatase 1B-Deficient Mice". Mol. Cell. Biol. 20 (15): 5479–89. doi:10.1128/MCB.20.15.5479-5489.2000. PMC 85999. PMID 10891488.
- ↑ Flint AJ, Tiganis T, Barford D, Tonks NK (March 1997). "Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases". Proc. Natl. Acad. Sci. U.S.A. 94 (5): 1680–5. doi:10.1073/pnas.94.5.1680. PMC 19976. PMID 9050838.
- ↑ Buckley DA, Cheng A, Kiely PA, Tremblay ML, O'Connor R (April 2002). "Regulation of Insulin-Like Growth Factor Type I (IGF-I) Receptor Kinase Activity by Protein Tyrosine Phosphatase 1B (PTP-1B) and Enhanced IGF-I-Mediated Suppression of Apoptosis and Motility in PTP-1B-Deficient Fibroblasts". Mol. Cell. Biol. 22 (7): 1998–2010. doi:10.1128/MCB.22.7.1998-2010.2002. PMC 133665. PMID 11884589.
- ↑ Heinonen KM, Dubé N, Bourdeau A, Lapp WS, Tremblay ML (February 2006). "Protein tyrosine phosphatase 1B negatively regulates macrophage development through CSF-1 signaling". Proc. Natl. Acad. Sci. U.S.A. 103 (8): 2776–81. doi:10.1073/pnas.0508563103. PMC 1413784. PMID 16477024.
- ↑ Zhu S, Bjorge JD, Fujita DJ (November 2007). "PTP1B contributes to the oncogenic properties of colon cancer cells through Src activation". Cancer Res. 67 (21): 10129–37. doi:10.1158/0008-5472.CAN-06-4338. PMID 17974954.
- ↑ 18.0 18.1 Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP et al. (December 2001). "TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B". J. Biol. Chem. 276 (51): 47771–4. doi:10.1074/jbc.C100583200 (inactive 2015-01-01). PMID 11694501.
- ↑ Zhang Z, Lin SY, Neel BG, Haimovich B (January 2006). "Phosphorylated alpha-actinin and protein-tyrosine phosphatase 1B coregulate the disassembly of the focal adhesion kinase x Src complex and promote cell migration". J. Biol. Chem. 281 (3): 1746–54. doi:10.1074/jbc.M509590200. PMID 16291744.
- ↑ 20.0 20.1 20.2 Liu F, Hill DE, Chernoff J (December 1996). "Direct binding of the proline-rich region of protein tyrosine phosphatase 1B to the Src homology 3 domain of p130(Cas)". J. Biol. Chem. 271 (49): 31290–5. doi:10.1074/jbc.271.49.31290. PMID 8940134.
- ↑ Dubé N, Cheng A, Tremblay ML (February 2004). "The role of protein tyrosine phosphatase 1B in Ras signaling". Proc. Natl. Acad. Sci. U.S.A. 101 (7): 1834–9. doi:10.1073/pnas.0304242101. PMC 357013. PMID 14766979.
- ↑ Balsamo J, Arregui C, Leung T, Lilien J (October 1998). "The Nonreceptor Protein Tyrosine Phosphatase PTP1B Binds to the Cytoplasmic Domain of N-Cadherin and Regulates the Cadherin–Actin Linkage". J. Cell Biol. 143 (2): 523–32. doi:10.1083/jcb.143.2.523. PMC 2132848. PMID 9786960.
- ↑ Stuible M, Dubé N, Tremblay ML (June 2008). "PTP1B regulates cortactin tyrosine phosphorylation by targeting Tyr446". J. Biol. Chem. 283 (23): 15740–6. doi:10.1074/jbc.M710534200. PMC 3259645. PMID 18387954.
- ↑ 24.0 24.1 24.2 Tonks NK (Jul 3, 2003). "PTP1B: from the sidelines to the front lines!". FEBS Letters 546 (1): 140–8. doi:10.1016/s0014-5793(03)00603-3. PMID 12829250.
- ↑ 25.0 25.1 Barford D, Flint AJ, Tonks NK (March 1994). "Crystal structure of human protein tyrosine phosphatase 1B". Science 263 (5152): 1397–404. doi:10.1126/science.8128219. PMID 8128219.
- ↑ Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A et al. (2004). "Protein tyrosine phosphatases in the human genome.". Cell 117 (6): 699–711. doi:10.1016/j.cell.2004.05.018. PMID 15186772.
- ↑ Mahadev K, Zilbering A, Zhu L, Goldstein BJ (2001). "Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase 1b in vivo and enhances the early insulin action cascade". J. Biol. Chem. 276 (24): 21938–42. doi:10.1074/jbc.C100109200. PMID 11297536.
- ↑ Lee SR, Kwon KS, Kim SR, Rhee SG (June 1998). "Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor". J. Biol. Chem. 273 (25): 15366–72. doi:10.1074/jbc.273.25.15366. PMID 9624118.
- ↑ Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T (October 1995). "Requirement for generation of H2O2 for platelet-derived growth factor signal transduction". Science 270 (5234): 296–9. doi:10.1126/science.270.5234.296. PMID 7569979.
- ↑ 30.0 30.1 Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK et al. (2003). "Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate.". Nature 423 (6941): 769–73. doi:10.1038/nature01680. PMID 12802338.
- ↑ 31.0 31.1 van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H (2003). "Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.". Nature 423 (6941): 773–7. doi:10.1038/nature01681. PMID 12802339.
- ↑ 32.0 32.1 Ravichandran LV, Chen H, Li Y, Quon MJ (October 2001). "Phosphorylation of PTP1B at Ser(50) by Akt impairs its ability to dephosphorylate the insulin receptor". Mol. Endocrinol. 15 (10): 1768–80. doi:10.1210/mend.15.10.0711. PMID 11579209.
- ↑ Sarmiento M, Puius YA, Vetter SW, Keng YF, Wu L, Zhao Y et al. (July 2000). "Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition". Biochemistry 39 (28): 8171–9. doi:10.1021/bi000319w. PMID 10889023.
- ↑ Zhang ZY, Walsh AB, Wu L, McNamara DJ, Dobrusin EM, Miller WT (March 1996). "Determinants of substrate recognition in the protein-tyrosine phosphatase, PTP1". J. Biol. Chem. 271 (10): 5386–92. doi:10.1074/jbc.271.10.5386. PMID 8621392.
- ↑ 35.0 35.1 Goldstein BJ, Bittner-Kowalczyk A, White MF, Harbeck M (February 2000). "Tyrosine dephosphorylation and deactivation of insulin receptor substrate-1 by protein-tyrosine phosphatase 1B. Possible facilitation by the formation of a ternary complex with the Grb2 adaptor protein". J. Biol. Chem. 275 (6): 4283–9. doi:10.1074/jbc.275.6.4283. PMID 10660596.
- ↑ Thareja S, Aggarwal S, Bhardwaj TR, Kumar M (2012). "Protein tyrosine phosphatase 1B inhibitors: a molecular level legitimate approach for the management of diabetes mellitus". Med Res Rev 32 (3): 459–517. doi:10.1002/med.20219. PMID 20814956.
- ↑ Tonks NK (2013). "Protein tyrosine pho sphatases — from housekeeping enzymes to master regulators of signal transduction". FEBS J. 280 (2): 346–78. doi:10.1111/febs.12077. PMC 3662559. PMID 23176256.
- ↑ Dubé N, Bourdeau A, Heinonen KM, Cheng A, Loy AL, Tremblay ML (2005). "Genetic ablation of protein tyrosine phosphatase 1B accelerates lymphomagenesis of p53-null mice through the regulation of B-cell development". Cancer Res. 65 (21): 10088–95. doi:10.1158/0008-5472.CAN-05-1353. PMID 16267035.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||