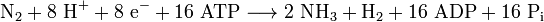Nitrogenase
| nitrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 1.18.6.1 | ||||||||
| CAS number | 9013-04-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Nitrogenases (EC 1.18.6.1EC 1.19.6.1) are enzymes used by some organisms to fix atmospheric nitrogen gas (N2). There is only one known family of enzymes that accomplishes this process. Whilst the equilibrium formation of ammonia from molecular hydrogen and nitrogen has an overall negative enthalpy of reaction (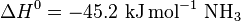 ), the activation energy is very high (
), the activation energy is very high (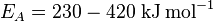 ).[1] Nitrogenase acts as a catalyst, reducing this energy barrier such that the reaction can take place at ambient temperatures.
).[1] Nitrogenase acts as a catalyst, reducing this energy barrier such that the reaction can take place at ambient temperatures.
Introduction
The enzymatic reduction of dinitrogen to ammonia requires both a reducing agent, such as ferredoxin or flavodoxin and an input of chemical energy, released from the hydrolysis of ATP, to overcome the activation energy barrier.[2] The enzyme is composed of the heterotetrameric MoFe protein that is transiently associated with the homodimeric Fe protein. Electrons for the reduction of nitrogen are supplied to nitrogenase when it associates with the reduced, nucleotide-bound homodimeric Fe protein. The heterocomplex undergoes cycles of association and disassociation to transfer one electron, which is the rate-limiting step in nitrogen reduction . ATP supplies the energy to drive the transfer of electrons from the Fe protein to the MoFe protein. The reduction potential of each electron transferred to the MoFe protein is sufficient to break one of dinitrogen's chemical bonds, though it has not yet been shown that exactly three cycles are sufficient to convert one molecule of N2 to ammonia. Nitrogenase ultimately bonds each atom of nitrogen to three hydrogen atoms to form ammonia (NH3), which is in turn bonded to glutamate to form glutamine. The nitrogenase reaction additionally produces molecular hydrogen as a side product.
The exact mechanism of catalysis is unknown due to the difficulty in obtaining crystals of nitrogen bound to nitrogenase. This is because the resting state of the MoFe protein does not bind nitrogen and also requires at least three electron transfers to perform catalysis. Nitrogenase is able to reduce acetylene, but is inhibited by carbon monoxide, which binds to the enzyme and thereby prevents binding of dinitrogen. Dinitrogen will prevent acetylene binding, but acetylene does not inhibit binding of dinitrogen and requires only one electron for reduction to ethylene.[3]
All nitrogenases have an iron and sulfur-containing cofactor that includes an iron-sulfur cluster at the active site. In most proteins, this Fe-S cluster also contains molybdenum, in which case the active site is referred to as FeMoco. In some case, molybdenum is replaced by a vanadium or iron.[4]
Due to the oxidative properties of oxygen, most nitrogenases are irreversibly inhibited by dioxygen, which degradatively oxidizes the Fe-S cofactors. This requires mechanisms for nitrogen fixers to protect nitrogenase from oxygen in vivo. Despite this problem, many use oxygen as a terminal electron acceptor for respiration. One known exception is the nitrogenase of Streptomyces thermoautotrophicus, which is unaffected by the presence of oxygen.[5] Although the ability of some nitrogen fixers such as Azotobacteraceae to employ an oxygen-labile nitrogenase under aerobic conditions has been attributed to a high metabolic rate, allowing oxygen reduction at the cell membrane, the effectiveness of such a mechanism has been questioned at oxygen concentrations above 70 µM (ambient concentration is 230 µM O2), as well as during additional nutrient limitations.[6]
The reaction that this enzyme performs is:
Nonspecific reactions
In addition to performing the reaction N≡N → 2 NH3, nitrogenase is also capable of catalyzing the following reactions:[8][9]
- HC≡CH → H2C=CH2
- N–=N+=O → N2 + H2O
- N=N=N– → N2 + NH3
- C≡N−
→ CH4, NH3, H3C–CH3, H2C=CH2 (CH3NH2) - N≡C–R → RCH3 + NH3
- C≡N–R → CH4, H3C–CH3, H2C=CH2, C3H8, C3H6, RNH2
- O=C=S → CO + H2S [10][11]
- O=C=O → CO + H2O [10]
- S=C=N– → H2S + HCN [11]
- O=C=N– → H2O + HCN, CO + NH3 [11]
Furthermore, dihydrogen functions as a competitive inhibitor,[12] carbon monoxide functions as a non-competitive inhibitor,[8][9] and carbon disulfide functions as a rapid-equilibrium inhibitor[10] of nitrogenase.
Vanadium nitrogenases have also been shown to catalzye the conversion of CO into alkanes through a reaction comparable to Fischer-Tropsch synthesis.
Organisms that synthesize nitrogenase
- Free-living diazotrophs, e.g.
- Green sulfur bacteria
- Cyanobacteria (with ex. Nostoc or without ex. Cyanothece[13] differentiated heterocysts)
- Azotobacteraceae
- Symbiotic diazotrophs, e.g.
Typically nitrogenases are coded by Nif genes.
Similarity to other proteins
The three subunits of nitrogenase exhibit significant sequence similarity to three subunits of the light-independent version of protochlorophyllide reductase that performs the conversion of protochlorophyllide to chlorophyll. This protein is present in gymnosperms, algae, and photosynthetic bacteria but has been lost by angiosperms during evolution.[14]
Separately, two of the nitrogenase subunits (NifD and NifD) have homologues in methanogens that do not fix nitrogen e.g. Methanocaldococcus jannaschii.[15] Little is understood about the function of these "class IV" nif genes,[16] though they occur in many methanogens. In M. jannaschii they are known to interact with each other and are constitutively expressed.[15]
Measurement of Nitrogenase Activity
As with many assays for enzyme activity, it is possible to estimate nitrogenase activity by measuring the rate of conversion of the substrate (N2) to the product (NH3). Since NH3 is involved in other reactions in the cell, it is often desirable to label the substrate with 15N to provide accounting or "mass balance" of the added substrate. A more common assay, the acetylene reduction assay or ARA, estimates the activity of nitrogenase by taking advantage of the ability of the enzyme to reduce acetylene gas to ethylene gas. These gases are easily quantified using gas chromatography.[17] Though first used in a laboratory setting to measure nitrogenase activity in extracts of Clostridium pasteurianum cells, ARA has been applied to a wide range of test systems, including field studies where other techniques are difficult to deploy. For example, ARA was used successfully to demonstrate that bacteria associated with rice roots undergo seasonal and diurnal rhythms in nitrogenase activity, which were apparently controlled by the plant.[18]
Unfortunately, the conversion of data from nitrogenase assays to actual moles of N2 reduced (particularly in the case of ARA), is not always straightforward and may either underestimate or overestimate the true rate for a variety of reasons. For example H2 competes with N2 but not acetylene for nitrogenase (leading to overestimates of nitrogenase by ARA). Bottle or chamber-based assays may produce negative impacts on microbial systems as a result of containment or disruption of the microenvironment through handling, leading to underestimation of nitrogenase. Despite these weaknesses, such assays are very useful in assessing relative rates or temporal patterns in nitrogenase activity.
See also
References
- Zumft WG, Mortenson LE (1975). "The nitrogen-fixing complex of bacteria". Biochim. Biophys. Acta. 416 (1): 1–52. doi:10.1016/0304-4173(75)90012-9. PMID 164247.
- ↑ Modak, J. M. (2002). "Haber Process for Ammonia Synthesis". Resonance 7: 69–77. doi:10.1007/bf02836187.
- ↑ Chi Chung, Lee; Markus W., Ribbe; Yilin, Hu (2014). "Chapter 7. Cleaving the N,N Triple Bond: The Transformation of Dinitrogen to Ammonia by Nitrogenases". In Peter M.H. Kroneck and Martha E. Sosa Torres. The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences 14. Springer. pp. 147–174. doi:10.1007/978-94-017-9269-1_6.
- ↑ Seefeldt, LC; Dance, IG; Dean, DR (2004). "Substrate interactions with nitrogenase: Fe versus Mo.". Biochemistry 43 (6): 1401–9. doi:10.1021/bi036038g.
- ↑ Robson, R. L.; Eady, R. R.; Richardson, T. H.; Miller, R. W.; Hawkins, M.; Postgate, J. R. (1986). "The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme". Nature (London) 322: 388–390. doi:10.1038/322388a0.
- ↑ Ribbe, M.; Gadkari, D.; Meyer, O. (1997). "N2 fixation by Streptomyces thermoautotrophicus involves a molybdenum-dinitrogenase and a manganese-superoxide oxidoreductase that couple N2 reduction to the oxidation of superoxide produced from O2 by a molybdenum-CO dehydrogenase". The Journal of biological chemistry 272 (42): 26627–26633. doi:10.1074/jbc.272.42.26627. PMID 9334244.
- ↑ Oelze, J (2000). "Respiratory protection of nitrogenase in Azotobacter species: Is a widely-held hypothesis unequivocally supported by experimental evidence?". FEMS Microbiol Rev 24 (4): 321–33. doi:10.1111/j.1574-6976.2000.tb00545.x.
- ↑ Igarashi, Robert; Seefeldt, Lance (2003). "Nitrogen fixation: the mechanism of the Mo-dependent nitrogenase". Critical reviews in biochemistry and molecular biology 38 (4): 351-384. doi:10.1080/10409230390242380.
- ↑ 8.0 8.1 Rivera-Ortiz, José M., and Burris, Robert H. (1975). "Interactions among substrates and inhibitors of nitrogenase". J Bacteriol 123 (2): 537–545. PMC 235759. PMID 1150625.
- ↑ 9.0 9.1 G. N. Schrauzer (2003). "Nonenzymatic Simulation of Nitrogenase Reactions and the Mechanism of Biological Nitrogen Fixation". Angewandte Chemie International Edition in English 14 (8): 514–522. doi:10.1002/anie.197505141. PMID 810048.
- ↑ 10.0 10.1 10.2 Lance C. Seefeldt, Madeline E. Rasche, Scott A. Ensign (1995). "Carbonyl sulfide and carbon dioxide as new substrates, and carbon disulfide as a new inhibitor, of nitrogenase". Biochemistry 34 (16): 5382–5389. doi:10.1021/bi00016a009. PMID 7727396.
- ↑ 11.0 11.1 11.2 Madeline E. Rasche and Lance C. Seefeldt (1997). "Reduction of Thiocyanate, Cyanate, and Carbon Disulfide by Nitrogenase: Kinetic Characterization and EPR Spectroscopic Analysis". Biochemistry 36 (28): 8574–8585. doi:10.1021/bi970217e. PMID 9214303.
- ↑ Joseph H. Guth, Robert H. Burris (1983). "Inhibition of nitrogenase-catalyzed ammonia formation by hydrogen". Biochemistry 22 (22): 5111–5122. doi:10.1021/bi00291a010. PMID 6360203.
- ↑ "Better living through cyanothece - unicellular diazotrophic cyanobacteria with highly versatile metabolic systems.". Adv Exp Med Biol 675: 275–90. 2010. doi:10.1007/978-1-4419-1528-3_16. PMID 20532747.
- ↑ Li, J.; Goldschmidt-Clermont, M.; Timko, M. P. (1993). "Chloroplast-Encoded chlB is Required for Light-Independent Protochlorophyllide Reductase Activity in Chlamydomonas reinhardtii". The Plant Cell Online 5 (12): 1817–1829. doi:10.1105/tpc.5.12.1817. PMC 160407. PMID 8305874.
- ↑ 15.0 15.1 Staples, C. R.; Lahiri, S.; Raymond, J.; Von Herbulis, L.; Mukhophadhyay, B.; Blankenship, R. E. (2007). "Expression and Association of Group IV Nitrogenase NifD and NifH Homologs in the Non-Nitrogen-Fixing Archaeon Methanocaldococcus jannaschii". Journal of Bacteriology 189 (20): 7392–7398. doi:10.1128/JB.00876-07. PMC 2168459. PMID 17660283.
- ↑ Raymond, J.; Siefert, J. L.; Staples, C. R.; Blankenship, R. E. (2003). "The Natural History of Nitrogen Fixation". Molecular Biology and Evolution 21 (3): 541–554. doi:10.1093/molbev/msh047. PMID 14694078.
- ↑ Dilworth, M.J. 1966. Acetylene reduction by nitrogen fixing preparations from Clostridium pasteurianum. Biochem. Biophys, Acta 127:285-29.
- ↑ Sims, GK and EP Dunigan. 1984. Diurnal and seasonal variations in nitrogenase activity (C2H2 reduction) of rice roots. Soil Biology and Biochemistry. 16:15-18.
| |||||||||||||||||||||||||||||||||||||