Neighbor-net
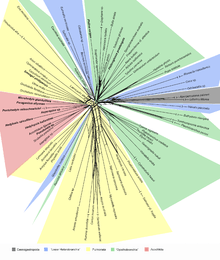
NeighborNet[1] is an algorithm for constructing phylogenetic networks which is loosely based on the neighbor joining algorithm. Like neighbor joining, the method takes a distance matrix as input, and works by agglomerating clusters. However, the NeighborNet algorithm can lead to collections of clusters which overlap and do not form a hierarchy, and are represented using a type of phylogenetic network called a split network. If the distance matrix satisfies the Kalmanson combinatorial conditions then Neighbor-net will return the corresponding circular ordering.[2][3] The method is implemented in the SplitsTree package.
Examples of the application of Neighbor-net can be found in virology[4] , horticulture,[5] dinosaur genetics,[6] comparative linguistics, and archaeology.[7]
References
- ↑ Bryant and Moulton : Neighbor-net, an agglomerative method for the construction of phylogenetic networks - Molecular Biology and Evolution 21 (2003)
- ↑ Bryant, D. and Moulton, V. and Spillner, A. 2007. Consistency of the NeighborNet algorithm. Algorithms in Molecular Biology, 2,8
- ↑ D. Levy and L. Pachter, The neighbor-net algorithm, Advances in Applied Mathematics, doi:10.1016/j.aam.2010.09.002.
- ↑ J. Schmidt-Chanasit, A. Bialonski, P. Heinemann, R. Ulrich, S. Gunther, H. Rabenau, and H. Doerr. A10-year molecular survey of herpes simplex virus type 1 in germany demonstrates a stable and high prevalence of genotypes a and b. Journal of Clinical Virology, 44(3):235 -- 237, 2009.
- ↑ Killian, B. Özkan, H., Deusch, O., Effgen, S., Brandolini, A., Kohl, J., Martin, W., Salamini, F. (2007). Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Molecular Biology and Evolution 24(1):217-227.
- ↑ M. Buckley and 27 others. Comment on``protein sequences from Mastodon and Tyrannosaurus Rex revealed by mass spectrometry". Science, 319(5859):33, 2008.
- ↑ S. Shennan. Pattern and process in cultural evolution. University of California Press, 2009.