Membrane computing
Membrane computing (or MC) is an area within computer science that seeks to discover new computational models from the study of biological cells, particularly of the cellular membranes. It is a sub-task of creating a cellular model.
Membrane computing deals with distributed and parallel computing models, processing multisets of symbol objects in a localized manner. Thus, evolution rules allow for evolving objects to be encapsulated into compartments defined by membranes. The communications between compartments and with the environment play an essential role in the processes. The various types of membrane systems are known as P systems after Gheorghe Păun who first conceived the model in 1998.[1]
An essential ingredient of a P system is its membrane structure, which can be a hierarchical arrangement of membranes, as in a cell, or a net of membranes (placed in the nodes of a graph), as in a tissue or a neural net. P systems are often depicted graphically with drawings.
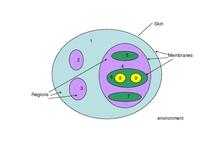
The intuition behind the notion of a membrane is a three-dimensional vesicle from biology. However the concept itself is more general, and a membrane is seen as a separator of two regions. The membrane provides for selective communication between the two regions. As per Gheorghe Păun, the separation is of the Euclidean space into a finite “inside” and an infinite “outside”. The selective communication is where the computing comes in.
Graphical representations may have numerous elements, according to the variation of the model that is being studied. For example, a rule may produce the special symbol δ, in which case the membrane that contains it is dissolved and all its contents move up in the region hierarchy.
The variety of suggestions from biology and the range of possibilities to define the architecture and the functioning of a membrane-based multiset processing device are practically endless. Indeed the membrane computing literature contains a very large number of models. Thus, MC is not merely a theory related to a specific model, it is a framework for devising compartmentalized models.
Chemicals are modeled by symbols, or alternatively by strings of symbols. The region, which is defined by a membrane, can contain other symbols or strings (collectively referred to as objects) or other membranes, so that a P system has exactly one outer membrane, called the skin membrane, and a hierarchical relationship governing all its membranes under the skin membrane.
If objects are symbols, then their multiplicity within a region matters; however multi-sets are also used in some string models. Regions have associated rules that define how objects are produced, consumed, passed to other regions and otherwise interact with one another. The nondeterministic maximally parallel application of rules throughout the system is a transition between system states, and a sequence of transitions is called a computation. Particular goals can be defined to signify a halting state, at which point the result of the computation would be the objects contained in a particular region. Alternatively the result may be made up of objects sent out of the skin membrane to the environment.
Many variant models have been studied, and interest has focused on proving computational universality for systems with a small number of membranes, for the purpose of solving NP-complete problems such as Boolean satisfiability (SAT) problems and the traveling salesman problem (TSP). The P systems may trade space and time complexities and less often use models to explain natural processes in living cells. The studies devise models that may at least theoretically be implemented on hardware. To date, the P systems are nearly all theoretical models that have never been reduced to practice, although a practical system is given in.[2]
See also
- Modeling biological systems
References
- ↑ Păun, Gheorghe. "Introduction to Membrane Computing".
- ↑ U.S. Patent 20,090,124,506