Meganuclease
Meganucleases are endodeoxyribonucleases characterized by a large recognition site (double-stranded DNA sequences of 12 to 40 base pairs); as a result this site generally occurs only once in any given genome. For example, the 18-base pair sequence recognized by the I-SceI meganuclease would on average require a genome twenty times the size of the human genome to be found once by chance (although sequences with a single mismatch occur about three times per human-sized genome). Meganucleases are therefore considered to be the most specific naturally occurring restriction enzymes.
Among meganucleases, the LAGLIDADG family of homing endonucleases has become a valuable tool for the study of genomes and genome engineering over the past fifteen years. Meganucleases are "molecular DNA scissors" that can be used to replace, eliminate or modify sequences in a highly targeted way. By modifying their recognition sequence through protein engineering, the targeted sequence can be changed. Meganucleases are used to modify all genome types, whether bacterial, plant or animal. They open up wide avenues for innovation, particularly in the field of human health, for example the elimination of viral genetic material or the "repair" of damaged genes using gene therapy.
Two main families
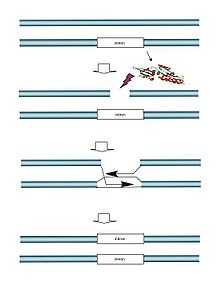
Meganucleases are found in a large number of organisms – Archaea or archaebacteria, bacteria, phages, fungi, yeast, algae and some plants. They can be expressed in different compartments of the cell – the nucleus, mitochondria or chloroplasts. Several hundred of these enzymes have been identified.
Meganucleases are mainly represented by two main enzyme families collectively known as homing endonucleases: intron endonucleases and intein endonucleases.
In nature, these proteins are coded by mobile genetic elements, introns or inteins. Introns propagate by intervening at a precise location in the DNA, where the expression of the meganuclease produces a break in the complementary intron- or intein-free allele. For inteins and group I introns, this break leads to the duplication of the intron or intein at the cutting site by means of the homologous recombination repair for double-stranded DNA breaks.
We know relatively little about the actual purpose of meganucleases. It is widely thought that the genetic material that codes for them functions as a parasitic element that uses the double-stranded DNA cell repair mechanisms to its own advantage as a means of multiplying and spreading, without damaging the genetic material of its host.
Homing endonucleases from the LAGLIDADG family
There are five families, or classes, of homing endonucleases.[1] The most widespread and best known is the LAGLIDADG family. It is mostly found in the mitochondria and chloroplasts of eukaryotic unicellular organisms.
Its name corresponds to an amino acid sequence (or motif) that is found, more or less conserved, in all the proteins of this family. These small proteins are also known for their compact and closely packed three-dimensional structures.
The best characterized endonucleases which are most widely used in research and genome engineering include I-SceI (discovered in the mitochondria of baker's yeast Saccharomyces cerevisiae), I-CreI (from the chloroplasts of the green algae Chlamydomonas reinhardtii) and I-DmoI (from the archaebacterium Desulfurococcus mobilis).
The best known LAGLIDADG endonucleases are homodimers (for example I-CreI, composed of two copies of the same protein domain) or internally symmetrical monomers (I-SceI). The DNA binding site, which contains the catalytic domain, is composed of two parts on either side of the cutting point. The half-binding sites can be extremely similar and bind to a palindromic or semi-palindromic DNA sequence (I-CreI), or they can be non-palindromic (I-SceI).
Meganucleases as tools for genome engineering
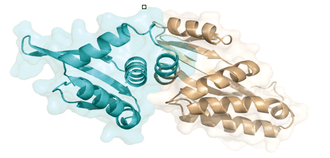
The high specificity of meganucleases gives them a high degree of precision and much lower cell toxicity than other naturally occurring restriction enzymes; they were identified in the 1990s as particularly promising tools for genome engineering.
However, the meganuclease-induced genetic recombinations that could be performed were limited by the repertoire of meganucleases available. Despite the existence of hundreds of meganucleases in nature, and the fact that each one is able to tolerate minor variations in its recognition site, the probability of finding a meganuclease able to cut a given gene at the desired location is extremely slim. Several research laboratories therefore soon began trying to engineer new meganucleases targeting the desired recognition sites.
The most advanced research and applications concern homing endonucleases from the LAGLIDADG family.
To create tailor-made meganucleases, two main approaches have been adopted:
- Modifying the specificity of existing meganucleases by introducing a small number of variations to the amino acid sequence and then selecting the functional proteins on variations of the natural recognition site.[3][4][5]
- A more radical option has been to exploit a property that plays an important role in meganucleases’ naturally high degree of diversification: the possibility of associating or fusing protein domains from different enzymes.[6][7] This option makes it possible to develop chimeric meganucleases with a new recognition site composed of a half-site of meganuclease A and a half-site of protein B. By fusing the protein domains of I-DmoI and I-CreI, two chimeric meganucleases have been created using this method: E-Drel and DmoCre.[8]
These two approaches can be combined to increase the possibility of creating new enzymes, while maintaining a high degree of efficacy and specificity. The scientists from Cellectis, a French biotechnology company, have developed a collection of over 20,000 protein domains from the homodimeric meganuclease I-CreI as well as from other meganucleases scaffolds.[2] They can be combined to form functional chimeric tailor-made heterodimers for research laboratories and for industrial purposes.
Precision Biosciences, an American biotechnology company, has developed a fully rational design process called Directed Nuclease Editor (DNE) which is capable of creating engineered meganucleases that target and modify a user-defined location in a genome.[9] In 2012 researchers at Bayer CropScience used DNE to incorporate a gene sequence into the DNA of cotton plants, targeting it precisely to a predetermined site.[10]
Probabilities
As stated in the opening paragraph, a meganuclease with an 18-base pair sequence would on average require a genome twenty times the size of the human genome to be found once by chance; the calculation is 418/3x109 = 22.9. However, very similar sequences are much more common, with frequency increasing quickly the more mismatches are permitted.
For example, a sequence which is identical in all but one base pair would on average occur by chance once every 417/18x3x109 = 0.32 human genome equivalents, or three times per human genome. A sequence which is identical in all but two base pairs would on average occur by chance once every 416/(18C2)x3x109 = 0.0094 human genome equivalents, or 107 times per human genome.
This is important because enzymes do not have perfect discrimination; a nuclease will still have some likelihood of acting even if the sequence does not match perfectly. So the activity of the nuclease on a sequence with one mismatch is less than the no-mismatch case, and activity is even less for two mismatches, but still not zero. Exclusion of these sequences, which are very similar but not identical, is still an important problem to be overcome in genome engineering.
See also
- Homing endonuclease
- homologous recombination
- I-CreI
- Protein engineering
- Protein design
- Genome editing with engineered nucleases
- Genome engineering
- Genetic engineering
References
- ↑ Stoddard, Barry L. (2006). "Homing endonuclease structure and function". Quarterly Reviews of Biophysics 38 (1): 49–95. doi:10.1017/S0033583505004063. PMID 16336743.
- ↑ 2.0 2.1 Grizot, S.; Epinat, J. C.; Thomas, S.; Duclert, A.; Rolland, S.; Paques, F.; Duchateau, P. (2009). "Generation of redesigned homing endonucleases comprising DNA-binding domains derived from two different scaffolds". Nucleic Acids Research 38 (6): 2006–18. doi:10.1093/nar/gkp1171. PMC 2847234. PMID 20026587.
- ↑ Seligman, L. M.; Chisholm, KM; Chevalier, BS; Chadsey, MS; Edwards, ST; Savage, JH; Veillet, AL (2002). "Mutations altering the cleavage specificity of a homing endonuclease". Nucleic Acids Research 30 (17): 3870–9. doi:10.1093/nar/gkf495. PMC 137417. PMID 12202772.
- ↑ Sussman, Django; Chadsey, Meg; Fauce, Steve; Engel, Alex; Bruett, Anna; Monnat, Ray; Stoddard, Barry L.; Seligman, Lenny M. (2004). "Isolation and Characterization of New Homing Endonuclease Specificities at Individual Target Site Positions". Journal of Molecular Biology 342 (1): 31–41. doi:10.1016/j.jmb.2004.07.031. PMID 15313605.
- ↑ Rosen, L. E.; Morrison, H. A.; Masri, S.; Brown, M. J.; Springstubb, B.; Sussman, D.; Stoddard, B. L.; Seligman, L. M. (2006). "Homing endonuclease I-CreI derivatives with novel DNA target specificities". Nucleic Acids Research 34 (17): 4791–800. doi:10.1093/nar/gkl645. PMC 1635285. PMID 16971456.
- ↑ Arnould, Sylvain; Chames, Patrick; Perez, Christophe; Lacroix, Emmanuel; Duclert, Aymeric; Epinat, Jean-Charles; Stricher, François; Petit, Anne-Sophie; Patin, Amélie (2006). "Engineering of Large Numbers of Highly Specific Homing Endonucleases that Induce Recombination on Novel DNA Targets". Journal of Molecular Biology 355 (3): 443–58. doi:10.1016/j.jmb.2005.10.065. PMID 16310802.
- ↑ Smith, J.; Grizot, S.; Arnould, S.; Duclert, A.; Epinat, J.-C.; Chames, P.; Prieto, J.; Redondo, P.; Blanco, F. J. (2006). "A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences". Nucleic Acids Research 34 (22): e149. doi:10.1093/nar/gkl720. PMC 1702487. PMID 17130168.
- ↑ Chevalier, Brett S.; Kortemme, Tanja; Chadsey, Meggen S.; Baker, David; Monnat, Raymond J.; Stoddard, Barry L. (2002). "Design, Activity, and Structure of a Highly Specific Artificial Endonuclease". Molecular Cell 10 (4): 895–905. doi:10.1016/S1097-2765(02)00690-1. PMID 12419232.
- ↑ Gao, Huirong; Smith, Jeff; Yang, Meizhu; Jones, Spencer; Djukanovic, Vesna; Nicholson, Michael G.; West, Ande; Bidney, Dennis; Falco, S. Carl (2010). "Heritable targeted mutagenesis in maize using a designed endonuclease". The Plant Journal 61 (1): 176–87. doi:10.1111/j.1365-313X.2009.04041.x. PMID 19811621.
- ↑ http://www.research.bayer.com/en/straight-into-the-cotton-genome.aspx[]