Malate dehydrogenase (decarboxylating)
| NAD-malic enzyme | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.1.1.39 | ||||||||
| CAS number | 9028-46-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
Malate dehydrogenase (decarboxylating) (EC 1.1.1.39) or NAD-malic enzyme (NAD-ME) is an enzyme that catalyzes the chemical reaction
- (S)-malate + NADP+
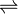 pyruvate + CO2 + NADPH
pyruvate + CO2 + NADPH
Thus, the two substrates of this enzyme are (S)-malate and NAD+, whereas its three products are pyruvate, CO2, and NADH. Malate is oxidized to pyruvate and CO2, and NAD+ is reduced to NADH.
This enzyme belongs to the family of oxidoreductases, to be specific, those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is (S)-malate:NAD+ oxidoreductase (decarboxylating). This enzyme participates in pyruvate metabolism and carbon fixation. NAD-malic enzyme is one of three decarboxylation enzymes used in the inorganic carbon concentrating mechanisms of C4 and CAM plants. The others are NADP-malic enzyme and PEP carboxykinase.[1][2]
Structural studies
As of late 2007, 7 structures have been solved for this class of enzymes, with PDB accession codes 1DO8, 1EFK, 1EFL, 1PJ2, 1PJ3, 1PJ4, and 1QR6.
References
- ↑ Kanai R, Edwards, GE (1999). "3. The Biochemistry of C4 Photosynthesis". In Sage RF, Monson RK. C4 Plant Biology. pp. 43–87. ISBN 0126144400.
- ↑ Christopher JT, Holtum JAM (1996). "Patterns of carbon partitioning in leaves of Crassulacean acid metabolism species during deacidification". Plant Physiol. 112 (1): 393–399. doi:10.1104/pp.112.1.393. PMC 157961. PMID 12226397.
- Saz HJ and Hubbard JA (1957). "The oxidative decarboxylation of malate by Ascaris lumbricoides". J. Biol. Chem. 225 (2): 921–933. PMID 13416294.