MTSL
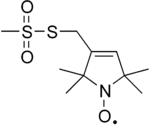 | |
| Names | |
|---|---|
| IUPAC name
S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate | |
| Other names
MTSL | |
| Identifiers | |
| 81213-52-7 | |
| ChemSpider | 117873 |
| |
| Jmol-3D images | Image |
| PubChem | 133628 |
| |
| Properties | |
| Molecular formula |
C10H18NO3S2 |
| Molar mass | 264.38 g·mol−1 |
| Except where noted otherwise, data is given for materials in their standard state (at 25 °C (77 °F), 100 kPa) | |
| | |
| Infobox references | |
MTSL (S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) is a chemical compound which can be used as a nitroxide (amine oxide) paramagnetic spin label in protein Electron paramagnetic resonance and nuclear magnetic resonance spectroscopy experiments. MTSL is attached via a disulfide bond to a cysteine residue, enabling site-directed spin labelling. Following attachment, which involves a Sulfinic acid (CH3SO2) leaving group,[1][2] the MTSL moiety will add 186.3 daltons to the mass of the protein or peptide to which it is attached. The cysteine can be introduced using site-directed mutagenesis, and hence most positions in a protein can be labelled.
In Nuclear magnetic resonance the introduction of the paramagnetic group increases the relaxation rate of nearby nuclei. This can be detected as peak broadening and loss of intensity in peaks corresponding to nearby nuclei. Hence proximity can be inferred for all nuclei, that are affected. A major advantage of this method over traditional methods for obtaining distance restraints in protein NMR is the increased length, as paramagnetic relaxation enhancement can detect distances up to 25 Å (2.5 nm) as opposed to about 6 Å (0.6 nm) using the nuclear Overhauser effect. Spin labelling with MTSL is frequently used in investigation of residual structure in intrinsically unstructured proteins.