Lignin peroxidase
| diarylpropane peroxidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.11.1.14 | ||||||||
| CAS number | 93792-13-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
Lignin is highly resistant to biodegradation and only higher fungi are capable of degrading the polymer via an oxidative process. This process has been studied extensively in the past twenty years, but the actual mechanism has not yet been fully elucidated. The complicated structure of the lignin polymer and major difficulties in analysis are responsible for the relatively slow progress
Lignin is found to be degraded by an enzyme lignin peroxidases produced by some fungi like Phanerochaete chrysosporium. The mechanism by which lignin peroxidase (Lip) interacts with the lignin polymer involves Veratryl alcohol (Valc). Which is a secondary metabolite of white rot fungi that acts as a cofactor for the enzyme.
In enzymology, a lignin peroxidase (EC 1.11.1.14) is an enzyme that catalyzes the chemical reaction
- 1,2-bis(3,4-dimethoxyphenyl)propane-1,3-diol + H2O2
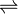 3,4-dimethoxybenzaldehyde + 1-(3,4-dimethoxyphenyl)ethane-1,2-diol + H2O
3,4-dimethoxybenzaldehyde + 1-(3,4-dimethoxyphenyl)ethane-1,2-diol + H2O
Thus, the two substrates of this enzyme are 1,2-bis(3,4-dimethoxyphenyl)propane-1,3-diol and H2O2, whereas its 3 products are 3,4-dimethoxybenzaldehyde, 1-(3,4-dimethoxyphenyl)ethane-1,2-diol, and H2O.
This enzyme belongs to the family of oxidoreductases, specifically those acting on a peroxide as acceptor (peroxidases) and can be included in the broad category of ligninases. The systematic name of this enzyme class is 1,2-bis(3,4-dimethoxyphenyl)propane-1,3-diol:hydrogen-peroxide oxidoreductase. Other names in common use include diarylpropane oxygenase, ligninase I, diarylpropane peroxidase, LiP, diarylpropane:oxygen,hydrogen-peroxide oxidoreductase (C-C-bond-cleaving). It employs one cofactor, heme.
Structural studies
As of late 2007, 3 structures have been solved for this class of enzymes, with PDB accession codes 1B80, 1B82, and 1B85.
References
- K.E.L. Eriksson, R.A. Blanchette and P. Ander (1990). "Microbial and Enzymatic Degradation of Wood and Wood Components,". Springer-Verlag.
- Hans E. Schoemaker and Klaus Piontek (1996). "On the interaction of lignin peroxidase with lignin,". Pure and Applied Chemistry 68 (11): 2089–96. doi:10.1351/pac199668112089.
- Paszczynski A, Huynh VB, Crawford R (1986). "Comparison of ligninase-I and peroxidase-M2 from the white-rot fungus Phanerochaete chrysosporium". Arch. Biochem. Biophys. 244 (2): 750–65. doi:10.1016/0003-9861(86)90644-2. PMID 3080953.
- Renganathan V, Miki K, Gold MH (1985). "Multiple molecular forms of diarylpropane oxygenase, an H2O2-requiring, lignin-degrading enzyme from Phanerochaete chrysosporium". Arch. Biochem. Biophys. 241 (1): 304–14. doi:10.1016/0003-9861(85)90387-X. PMID 4026322.
- Tien M and Kirk TT (1984). "Lignin-degrading enzyme from Phanerochaete chrysosporium purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase". Proc. Natl. Acad. Sci. USA 81 (8): 2280–2284. doi:10.1073/pnas.81.8.2280.
- Doyle WA, Blodig W, Veitch NC, Piontek K, Smith AT (1998). "Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis". Biochemistry. 37 (43): 15097–105. doi:10.1021/bi981633h. PMID 9790672.
- Wariishi H, Marquez L, Dunford HB, Gold MH (1990). "Lignin peroxidase compounds II and III. Spectral and kinetic characterization of reactions with peroxides". J. Biol. Chem. 265 (19): 11137–42. PMID 2162833.
- Cai DY, Tien M (1990). "Characterization of the oxycomplex of lignin peroxidases from Phanerochaete chrysosporium: equilibrium and kinetics studies". Biochemistry. 29 (8): 2085–91. doi:10.1021/bi00460a018. PMID 2328240.
- Tien M, Tu CP (1987). "Cloning and sequencing of a cDNA for a ligninase from Phanerochaete chrysosporium". Nature. 326 (6112): 520–3. doi:10.1038/326520a0. PMID 3561490.
- Renganathan V, Miki K, Gold MH (1986). "Role of molecular oxygen in lignin peroxidase reactions". Arch. Biochem. Biophys. 246 (1): 155–61. doi:10.1016/0003-9861(86)90459-5. PMID 3754412.
- Kersten PJ, Tien M, Kalyanaraman B, Kirk TK (1985). "The ligninase of Phanerochaete chrysosporium generates cation radicals from methoxybenzenes". J. Biol. Chem. 260 (5): 2609–12. PMID 2982828.
- Kirk TK, Farrell RL (1987). "Enzymatic "combustion": the microbial degradation of lignin". Annu. Rev. Microbiol. 41: 465–505. doi:10.1146/annurev.mi.41.100187.002341. PMID 3318677.