Isobaric labeling
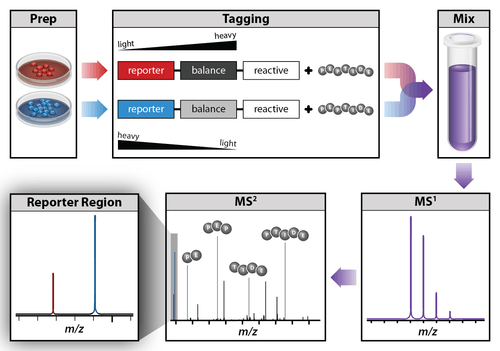
Isobaric labeling is a mass spectrometry strategy used in quantitative proteomics. Peptides or proteins are labeled with various chemical groups that are (at least nominally) isobaric, or the same in mass, but which fragment during tandem mass spectrometry to yield reporter ions of different mass. In a typical bottom-up proteomics workflow, proteins are enzymatically digested by a protease to produce peptides, which are then labeled with different isobaric tags. The samples are mixed in equal ratios. During a liquid chromatography-mass spectrometry analysis, the peptides are fragmented to produce sequence-specific product ions, which help to determine the peptide sequence, as well as the reporter tags, whose abundances reflect the relative ratio of the peptide in the samples that were combined.
Availability
There are two types of isobaric tags commercially available: tandem mass tags (TMT)[1] and isobaric tags for relative and absolute quantitation (iTRAQ).[2] TMT is available in duplex and 6-plex[3] forms, while iTRAQ is available in 4-plex and 8-plex[4] forms.
See also
- Isotope-coded affinity tag (ICAT)
- Stable isotope labeling with amino acids in cell culture (SILAC)
- Label-free quantitation
References
- ↑ Thompson A, Schäfer J, Kuhn K et al. (2003). "Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS". Anal. Chem. 75 (8): 1895–904. doi:10.1021/ac0262560. PMID 12713048.
- ↑ Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004). "Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents". Mol. Cell. Proteomics 3 (12): 1154–69. doi:10.1074/mcp.M400129-MCP200. PMID 15385600.
- ↑ Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC (2008). "Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags". Anal. Chem. 80 (8): 2921–31. doi:10.1021/ac702422x. PMID 18312001.
- ↑ Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH (2007). "8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease". Proteomics 7 (20): 3651–60. doi:10.1002/pmic.200700316. PMC 3594777. PMID 17880003.
Further reading
- Casado-Vela, Juan (2010). "iTRAQ-based quantitative analysis of protein mixtures with large fold change and dynamic range.". Proteomics 10 (2): 343–347. doi:10.1002/pmic.200900509. PMID 20029838.
| ||||||||||