Histidine—tRNA ligase
| histidine-tRNA ligase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 6.1.1.21 | ||||||||
| CAS number | 9068-78-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
In enzymology, a histidine-tRNA ligase (EC 6.1.1.21) is an enzyme that catalyzes the chemical reaction
- ATP + L-histidine + tRNAHis
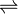 AMP + diphosphate + L-histidyl-tRNAHis
AMP + diphosphate + L-histidyl-tRNAHis
The 3 substrates of this enzyme are ATP, L-histidine, and tRNA(His), whereas its 3 products are AMP, diphosphate, and L-histidyl-tRNA(His).
This enzyme belongs to the family of ligases, to be specific those forming carbon-oxygen bonds in aminoacyl-tRNA and related compounds. The systematic name of this enzyme class is L-histidine:tRNAHis ligase (AMP-forming). Other names in common use include histidyl-tRNA synthetase, histidyl-transfer ribonucleate synthetase, and histidine translase. This enzyme participates in histidine metabolism and aminoacyl-trna biosynthesis.
Structural studies
As of late 2007, 9 structures have been solved for this class of enzymes, with PDB accession codes 1ADJ, 1ADY, 1H4V, 1HTT, 1KMM, 1KMN, 1QE0, 1WU7, and 1X59.
See also
- Anti-Jo1
References
- von Tigerstrom M, Tener GM (1967). "Histidyl transfer ribonucleic acid synthetase from bakers' yeast". Can. J. Biochem. 45 (7): 1067–74. doi:10.1139/o67-123. PMID 6035970.
| |||||||||||||||||||||