HLA-DQ8
| HLA-DQ8 (MHC Class II, DQ cell surface antigen) | |||
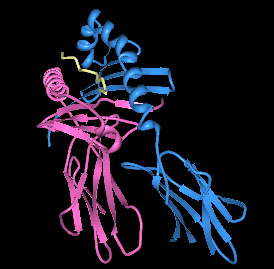 | |||
| Rendering of HLA-DQ8 with immundoinant insulin peptide in the binding pocket. | |||
| Cis-haplotype | Haplotype | ||
| isoform, | subtype | DQA1 | DQB1 |
| DQ α3β8 | DQ8.1 | *0301 | *0302 |
| DQ α3β8 | DQ8.1v | *0302 | *0302 |
| rare haplotypes | |||
| DQ α4β8 | DQ8.4 | *0401 | *0302 |
| DQ α5β8 | DQ8.5 | *0503 | *0302 |
HLA-DQ8 (DQ8) is a human leukocyte antigen serotype within the HLA-DQ (DQ) serotype group. DQ8 is a split antigen of the DQ3 broad antigen. DQ8 is determined by the antibody recognition of β8 and this generally detects the gene product of DQB1*0302.
DQ8 is commonly linked to autoimmune disease in the human population. DQ8 is the second most predominant isoform linked to coeliac disease and the DQ most linked to juvenile diabetes. DQ8 increases the risk for rheumatoid arthritis and is linked to the primary risk locus for RA, HLA-DR4. DR4 also plays an important role in juvenile diabetes. While the DQ8.1 haplotype is associated with disease, there is no known association with the DQB1*0305, DQ8.4 or DQ8.5 haplotypes (see infobox) with autoimmune disease; however, this may be the result of lack of study in populations that carry these and the very low frequency.
DQ8.1 also differs from other HLA in population frequencies. Typically for MHC Class II antigens in humans, haplotype frequencies do not exceed 40%. For example, in the US the highest haplotype frequency, the haplotype that encoded DQ6.2, is around 15%, this translates into phenotype frequencies of less than 30%. Atypically haplotype frequencies exceed 40%.
For DQ8 the highest haplotype frequencies approach 80% in parts of Central and South America and the phenotype frequencies approach 90%. This is the highest phenotype frequency observed for any DR or DQ phenotype in the human population by a wide margin.
Serology
| DQB1* | DQ8 | DQ3 | DQ7 | Sample |
| allele | % | % | % | size (N) |
| *0302 | 66 | 23 | 4 | 6687 |
| *0304 | 8 | 35 | 40 | 111 |
| *0305 | 34 | 30 | 70 | |
| Red indicates the level of 'false' reaction in non-DQ8 serotypes | ||||
| Alleles link-out to IMGT/HLA Databease at EBI | ||||
| freq | ||
| ref. | Population | (%) |
| [2] | Guatemala Mayans | 48.1 |
| Mexico N.Leon Mestizos | 22.5 | |
| USA South Texas Hispanics | 20.6 | |
| Sweden | 18.7 | |
| Russia Siberia Negidal | 18.6 | |
| Russia Murmansk Saomi | 18.5 | |
| Jordan Amman | 17.8 | |
| Samoa | 17.2 | |
| England Caucasoid | 16.4 | |
| Finland | 15.7 | |
| France | 14.5 | |
| Japan Central | 10.8 | |
| Greece Crete | 9.2 | |
| Spain Basque Arratia Valley | 6.7 | |
| Algeria Oran | 6.6 | |
| China Beijing and Xian | 6.1 | |
| Ethiopia Amhara | 5.6 | |
| USA SE African American | 4.9 | |
| USA Alaska Yupik Natives | 3.8 | |
| India North Hindus | 3.0 | |
| Zimbabwe Harare Shona | 2.2 | |
| Russia Siberia Eskimos | 0.9 | |
| Rwanda Kigali Hutu and Tutsi | 0.5 | |
| PNG Eastern Highlands Goroka | 0.0 | |
| Tunisia Jerba Berber | 0.0 |
Although false reaction with DQB1*0302 is low, the efficiency of the positive reaction is not good and there is a risk of false detection of DQB1*0305 which could create incompatibility. For disease diagnosis and confirmation, there is no known association of DQB1*0305 with either coeliac or autoimmune diabetes. Therefore it is prudent to use high resolution DQB1 typing for DQ8.
Alleles
| DQ | DQ | DQ | Freq | |||||
|---|---|---|---|---|---|---|---|---|
| Serotype | cis-isoform | Subtype | A1 | B1 | %[3] | rank | ||
| DQ8 | α3-β8 | 8.1 | 0301 | 0302 | 9. | 62 | 6th | |
| 0302♠ | 0302 | 0. | 93 | |||||
| ♠DQA1*0302 & *0303 not resolved | ||||||||
DQ8 is determined by the antibody recognition of β8 and is complicated by the fact that DQ8 recognizes some HLA-DQB1*03 encoded isoforms well, partially or not well at all (See serology) DQ β3.2 and β3.5 are best recognized as DQ8. These split antigens are the allele products of the DQB1*0302 and DQB1*0305, respectively.
DQB1*0302
| freq | ||
| ref. | Population | (%) |
| [2] | Lebanon Kafar Zubian | 5.9 |
| Lebanon Niha el Shouff | 2.5 | |
| Lakota Sioux | 2.1 | |
| Tunisia | 2.0 | |
| Lebanon Yuhmur | 1.8 | |
| Guatemala Mayans | 1.5 | |
| Morocco | 1.0 | |
| Pakistan | 1.0 | |
| Saudi Arabia Guraiat and Hail | 0.8 | |
| China Lijiang Naxi | 0.7 | |
| Russia Chuvash | 0.6 | |
| Tunisia Matmata Berber | 0.6 | |
| Czech Republic | 0.5 | |
| India Delhi | 0.5 |
DQB1*0302 and is found most often in the haplotype DQA1*0301:DQB1*0302, about 10% of the time it is found in the haplotype DQA1*0302:DQB1*0302. DQB1*0302 are almost always linked to DR4, DRB1*0401, *0402, and *0404 in caucasians. The first and third DRB1 are most strongly associated with rheumatoid arthritis.
DQB1*0305
DQB1*0305 gene product reacts slightly more intensely with DQ8 than DQ7 its generally rare in Europe and North America, except in a few indigeonous populations. Levels of DQB1*0305 are probably higher given earlier tests did not discriminate well between different *03.
Haplotypes
DQ8 β-chains combine with α-chains, encoded by genetically linked HLA-DQA1 alleles, to form the cis-haplotype isoforms. There is only one common cis-isoform of DQ8 because the linked DQA1*03 alleles(2) occur over the majority of the population, DQ8.1 is the overwhelming majority cis-isoform of DQ8. A rare haplotype DQA1*0503:DQB1*0302 is detected below 1% of all DQ8 haplotypes in Asia and Mesoamerica. Another rarer haplotype, DQA1*0401:DQB1*0302
DQ8.1
| Reference | DQA1 | DQB1 | Estimated | |
|---|---|---|---|---|
| Population | *03 | *0302 | DQ8.1 | |
| [4] | Lacandon Mayan (Mexico) |
79.0 | 77.9 | 77.9 |
| [5] | Perija-Yucpa (Venezuela) |
74.0 | 75.0 | 74.9 |
| [6] | Mayan (Guatemala) |
48.1 | 47.6 | |
| [7] | Mazatecans (Mexico) |
48.5 | 48.5 | 47.5 |
| [8] | Lamas (Peru) |
45.2 | 44.7 | |
| [9] | Dakota Sioux (S. Dakota) |
52.1 | 45.0 | 44.5 |
| [10] | Mixtec (Oaxaca, Mexico) |
40.0 | 35.9 | 35.4 |
| [11] | Lakota Sioux (S. Dakota) |
25.7 | 25.5 | |
| [12] | Terena (Brazil) |
17.5 | 17.0 | |
| [13] | Cauc., San Antonio (USA) |
11.7 | 11.7 | |
| [14] | Caucasian (USA) |
18.5 | 10.5 | 10.5 |
| [15] | African American (SE. USA) |
4.9 | 4.5 | |
| [16] | Tlinglet (Alaska, USA) |
14.0 | 8.5 | 8.5 |
| [17] | Eskimo (Alaska, USA) |
3.8 | 3.8 | |
| [18] | Canoncito Navajo (NM, USA) |
6.3 | 3.5 | 3.5 |
| [19] | Eskimo (E. Greenland) |
0.0 | 0.0 | 0.0 |
DQA1*0301:DQB1*0302 (DQ8.1) is the most common DQ8 subtype representing over 98% of the DQ8 bearing population. Infrequently, DQA1*0302:DQB1*0302, but this substitution of the alpha chain, DQA1**0301 versus *0302, is outside the binding cleft and appears not to alter DQ8 function. DQ8.1 is found almost ubiquitously in every human regional population, but because of its unique distribution it becomes an object of molecular anthropology. There are 3 places where haplotype frequency is elevated, Central and South America, NE Pacific Rim, and Northern Europe.
High frequencies in the Americas
The global node for DQ8 is in Central America and northern South America where it reaches the highest frequency for any single DQ serotype, close to 90% phenotype frequency (77% haplotype frequency), and is at relatively high frequency in the indigenous North American population, and the coastal regions of the Gulf of Mexico and up the Mississippi Valley. The high freqeuncy of DQ8 in South America's northeastern regions[5] and low frequency in Indigenous Americans of more recent Asian ancestry [16][19] or Siberian origin[20] suggest that DQ8 was at high frequency in the earliest Amerinds. The pattern of distribution is consistent with recent mtDNA results suggesting the first migrants to the New World settled in the lowland coastal regions, river valleys and moved slowly inland, subsequent settlers moved into the highland regions. DQ8 and DQ2.5 have many analogous functional similarities, and this first settler bias may be a reason for the similarity. Studies on Epstein Barr Virus[21] and other proteins suggest both proteins are acidic (meaning peptides with increased negative charge) peptide presenters (see DQ8 for an illustration of the presentation process) and may have been adaptive for certain hunting and gathering lifestyles, possibly coastal foragers.
| Reference | DQA1 | DQB1 | Estimated | |
|---|---|---|---|---|
| Population | *03 | *0302 | DQ8.1 | |
| [20] | Nivkhi (NNE. Sakalin I.) |
0.0 | 0 | |
| [22] | Zorastra, Yadz (Iran) |
20.8 | 0.8 | 0.8 |
| [23] | Khoton (Mongolia) |
24.4 | 1.2 | 1.2 |
| [24] | Madang (Papua New Guinea) |
1.5 | 1.5 | 1.5 |
| [25] | Yao (China) |
2.6 | 2.6 | |
| [25] | Naxi (Lijiang, China) |
2.7 | 2.7 | |
| [26] | Ainu (Japan) |
3.0 | 3.0 | |
| [27] | Shandong Han (China) |
3.1 | 3.1 | |
| [28] | Khalkha (Mongolia) |
22.3 | 6.1 | 6.1 |
| [29] | Thais | 21.5 | 7.4 | 7.4 |
| [30] | Ket, Yenisey (Siberia) |
14.7 | 8.4 | 8.4 |
| [31] | South Korea | 10.3 | 10.3 | |
| [32] | Japanese | 10.8 | 10.8 | |
| [33] | Nganasan (Siberia) |
39.6 | 11.4 | 11.4 |
| [34] | Uygars, Ürümqi (China) |
21.9 | 11.4 | 11.4 |
| [35] | Kazahk | 11.4 | 11.4 | |
| [36] | Negidal (Siberia) |
50.0 | 18.6 | 18.6 |
| [37] | Cook Isl. (Pacific) |
25.0 | 25.0 | |
Abundance in Asia
Hiatus of DQ8 in the NE Siberian Arctic, Elevated Levels in Amur Region and Eastern Turks
The levels of DQ8 in SW to West Pacific Rim are at variable haplotype frequencies, from 2 to 30%, and level off around 10% for Ryūkyūan, Japanese, Koreans, Amur Regions and in the NW Pacific Rim drop to less than 1% in the Nivkhi. There is a modern hiatus of DQ8 in the Alaska-Eastern Siberian region and it is unclear whether this is due to replacement, selection, or the mode in which first Americans arrived (i.e. strictly maritime route). The DR types associated with DQ8 are DRB1*0403, *0404, *0406, *0407, *0408, and *0401 is split between many DQA1:B1 haplotypes. DQB1*0405 is commonly associated with DQA1*0303:DQB1*04 and so it is not included in DRB1*0401 in high resolution assessments. The Cook Island DQ8 had only one associated DR haplotype suggesting diversity limiting introduction into the region, either via the TW-(Japan/Korea/China) route or through the west, for example the Bunun have high DRB1*0403. The majority of DRB1*04 appear to have redistributed from eastern Asia from an unknown source, possibly in Central Asia or India. The distribution can be compared with Native Groups such as South Americans. Three groups with high levels, the Kogui, Sikuni, and Yucpa, have about 75% DQ8, the dominant DRB1* allele in 2 of 3 is the *0411 (N. China = 0), but *0407 (Ryūkyū, Japanese, Mansi-Eastern Ural, Naxi Chinese) and *0403 (Nganasan, Buryat, Negidal, Tunisians, Ryūkyū, Korea, Ainu) are also found. In North America DRB1*0404 and *0407 are more common than *0403 and, in the Lakota Souix, B1*0411 is rare. The DRB1*0404-DQ8 haplotype is more common in North Western Asia, and Northern Europe.
| Reference | DQA1 | DQB1 | Estimated | |
|---|---|---|---|---|
| Population | *03 | *0302 | DQ8.1 | |
| [38] | Nenets (N. Russia) |
40.9 | 20.9 | 20.0 |
| [38] | Murmansk Saomi (Russia) |
38.3 | 18.5 | 17.2 |
| [39] | Gaza (Israel/Palestine) |
17.6 | 17.0 | |
| [40] | Arkhange Pomors (Russia) |
22.6 | 17.1 | 16.4 |
| [41] | Swedish | 24.2 | 18.7 | 16.1 |
| [41] | Caucasian (England) |
23.7 | 16.4 | 15.9 |
| [41] | Finland | 15.7 | 15.2 | |
| [42] | France's CEPH | 21.3 | 14.5 | 13.5 |
| [41] | Dane | 20.9 | 13.2 | 13.2 |
| [43] | Dutch | 11.2 | 11.0 | |
| Irish | 10.6 | |||
| [44] | NW Slavic (Russia) |
16.0 | 11.0 | 10.5 |
| [45] | German | 10.5 | 9.9 | |
| [46] | Russian | 8.9 | 7.2 | |
| [47] | Cantabarian (Spain) |
19.3 | 8.4 | 7.0 |
| [45] | Spanish | 8.9 | 6.5 | |
| [45] | Sardinian | 4.9 | 4.9 | |
| [45] | Italian | 4.6 | 4.5 | |
| [48] | United Arab Emirates | 0.83 | 0.8 | |
| [49] | Jordon | 24.1 | 17.9 | 17.9 |
| Africa | ||||
| [45] | !kung | 36.7 | 36.7 | |
| [45] | Khoi | 14.9 | 13.9 | |
| [50] | Oromo | 17.5 | 9.0 | 9.0 |
| [51] | Morocco | 17.2 | 8.9 | 8.6 |
| [52] | Tunisia | 10.31 | 10.0 | 8.0 |
| [50] | Amhara | 11.2 | 5.6 | 5.6 |
| [53] | Aka pygmies | 3.2 | 3.0 | |
| [45] | Negroid (N. Africa) |
3.1 | 2.7 | |
| [54] | Cameron | 11.5 | 1.5 | 1.2 |
| [53] | Bantu Congo | 0.6 | 0.5 | |
| [55] | Gabonese | 7.5 | 0.5 | |
| [56] | Bubi | 17.5 | 0.0 | 0.0 |
High levels of DQ8 in Northern Europe
DQ8 is also abundant also in Northern Europe and is found at high frequencies in the German-Scandinavian-Uralic population north of Switzerland. HLA A-B haplotypes suggest that a migration from people east of the Urals is responsible for DQ8, possibly from as far east as the West Pacific Rim. The high level of DQ8 and DQ2.5 is something of great interest for DQ mediated diseases of Scandinavia and Northern Europe. DQ8 is also found in Iberia and places were east to west gene flow by other genetic markers cannot be substantiated, and the levels within the African or middle eastern population are possible sources, Iberia has considerable A1/B1 equilibration suggesting independent sources from Africa.
Global spread of DQ8
DQ8 along with a few other haplotypes appears to be split NW/SE in Eurasia and with the evidence for DQ2.5 and other haplotypes suggest an ancient Central Asian population was displaced by a more recent African migration. There are many common markers found in France, Germans, Danes, Swedes, Tibetans, Amur River, Japanese and Koreans that are potential indicators of this bilateral spread. The DQ8 haplotypes is found at high frequencies in the !kung, albeit one expects more DQ8 in Austronesia it is ubiquitously spread if at some times low frequencies, other times higher frequencies (Thai). The path of DQ8 spread to the New World is enigmatic, certainly Japan and Amur River are potent sources, but other displaced populations cannot be ruled out. If the mode of travel was through the Beringia corridor as proposed by archaeologist, the very low frequency of DQ8 at present is a very unusual find with regard to evidence for complete displacement elsewhere in the World. Markers that are shared between Japanese, TW-aboriginals tend to decline in frequency as one approaches Siberia, mtDNA markers decline in the Kuril chain. During the Jōmon period of Japan it appears there would have been displacement by Ninhvet/Ainu ancestors and depression of DQ8 throughout northern Japan, but the decline throughout the region is somewhat inexplicable outside of a catastrophic climate event between the settling of the New World and the current time.
An alternative model is that there were multiple sources of DQ8 in the peopling of NE Asia, some sources were from central Asia and some from the indochinese region, some of the DQ8 found in NW Eurasia could be from an admixture of West pacific Rim and Central Asian sources, and were displaced from the more central regions but not from the more Eastern regions.
DQ8 and selection
Like DQ2.5, DQ8 might have been under selection for maritime, coastal foraging peoples and in particular for peoples adapted to the climate/habitat situation on the northern end of the habitable west pacific rim at the Last Glacial Maximum. Triticeae cultivation may apply negative selection on DQ8. While there were numerous members of Triticeae species similar to Mid-Eastern wild Triticeae in the Americas, and a great number of domesticated plants in the new world, no single species of Triticeae appears to have been domesticated in the New World, and no clear examples in closely related tribes of grasses. Among new world grass species in post Columbian times, one species of Elymus has been domesticated for human consumption and another as a pastoral cultivar. This could be interpreted in 2 ways. First, that levels of DQ8, negatively, inhibited the domestication of Triticeae strains. Second, that the absence of such cultivars more suitable than already developed cultivars allowed DQ8 to rise or remain high, while DQ2.5 levels in NW under much longer term selection have fallen, or a little of both. Most of American cultivars were domesticated south of the Rio Grande (exceptions are Caddo rice and Texas varigated squash, etc.). Wheat, particularly barley and rye, are preferential cultivars in cooler climate, whereas Zea is more adaptive in tropical climates and some cultivars are relatively drought-tolerant, Zea however lacks certain amino acids that must be supplemented by other foods to prevent malnutrition. The proximity of neolithization to the Equator in the New World may have much to do with the unapparent negative selection of DQ8 relative to the neolithization of Western Eurasia.
DQ8 and disease
In Europe, DQ8 is associated with juvenile diabetes and coeliac disease. The highest risk factor for type 1 diabetes is the HLA DQ8/DQ2.5 phenotype. In parts of eastern Scandinavia both DQ2.5 and DQ8 are high increases frequencies of late onset Type I and ambiguous Type I/II diabetes. DQ8 is also found in many indigenous peoples of Asia, it was detected early on in the Bedoin population of Arabia where DQ2.5 is frequently absent, and in these instances DQ8 is solely associated HLA in Coeliac Disease.
In the United States, however there appears to be shift in autoimmune disease risk for immigrants from Mexico. Increased immunoreactivity of Hispanics in Houston appear to be associated with DR4-DQ8. The haplotype may incur the highest risk for rheumatoid arthritis.
In Japan DQ3 (DQ7, DQ8, DQ9) is associated with myasthenia gravis in the early onset female population, though it does not appear DQ8 has the greater role, there are similarities between myasthenia gravis in Japan and that detected in the Houston Hispanic population, with DQ8 associated with younger females relative to the associations of all other HLA DQ types. Coeliac disease is on the rise in Japan, and it is clear that dietary shifts are the reason, but, also there is no DQ2.5 in Japan while Dq8 levels are moderate.
The DR4 linkage
Many disease associated with DQ8 have dual linkage with DR4, and certain DR4 (*0405) have independent and dependent risk association with DQ8, for example with juvenile diabetes.[57][58]
References
- ↑ Allele Query Form IMGT/HLA - European Bioinformatics Institute
- ↑ 2.0 2.1 Middleton D, Menchaca L, Rood H, Komerofsky R (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens 61 (5): 403–7. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
- ↑ Klitz W, Maiers M, Spellman S et al. (October 2003). "New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans". Tissue Antigens 62 (4): 296–307. doi:10.1034/j.1399-0039.2003.00103.x. PMID 12974796.
- ↑ "Review Population Study
(Proceedings of IHW Workshop, Seattle 2002) - Mexico Mayan Lacandon". Allele frequencies in worldwide population. Retrieved 2008-08-17. - ↑ 5.0 5.1 Layrisse Z, Guedez Y, Domínguez E, Paz N, Montagnani S, Matos M, Herrera F, Ogando V, Balbas O, Rodríguez-Larralde A (2001). "Extended HLA haplotypes in a Carib Amerindian population: the Yucpa of the Perija Range.". Hum Immunol 62 (9): 992–1000. doi:10.1016/S0198-8859(01)00297-X. PMID 11543901.
- ↑ Gómez-Casado E, Martínez-Laso J, Moscoso J, Zamora J, Martin-Villa M, Perez-Blas M, Lopez-Santalla M, Lucas Gramajo P, Silvera C, Lowy E, Arnaiz-Villena A (2003). "Origin of Mayans according to HLA genes and the uniqueness of Amerindians.". Tissue Antigens 61 (6): 425–36. doi:10.1034/j.1399-0039.2003.00040.x. PMID 12823766.
- ↑ Arnaiz-Villena A, Vargas-Alarcón G, Granados J, Gómez-Casado E, Longas J, Gonzales-Hevilla M, Zuñiga J, Salgado N, Hernández-Pacheco G, Guillen J, Martinez-Laso J (2000). "HLA genes in Mexican Mazatecans, the peopling of the Americas and the uniqueness of Amerindians.". Tissue Antigens 56 (5): 405–16. doi:10.1034/j.1399-0039.2000.560503.x. PMID 11144288.
- ↑ Moscoso J, Seclen S, Serrano-Vela J, Villena A, Martinez-Laso J, Zamora J, Moreno A, Ira-Cachafeiro J, Arnaiz-Villena A (2006). "HLA genes in Lamas Peruvian-Amazonian Amerindians.". Mol Immunol 43 (11): 1881–9. doi:10.1016/j.molimm.2005.10.013. PMID 16337001.
- ↑ "Review Population Study
(Proceedings of IHW Workshop, Seattle 2002) - USA South Dakota Sioux". Allele frequencies in worldwide population. Retrieved 2008-08-17. - ↑ Hollenbach J, Thomson G, Cao K, Fernandez-Vina M, Erlich H, Bugawan T, Winkler C, Winter M, Klitz W (2001). "HLA diversity, differentiation, and haplotype evolution in Mesoamerican Natives.". Hum Immunol 62 (4): 378–90. doi:10.1016/S0198-8859(01)00212-9. PMID 11295471.
- ↑ Leffell M, Fallin M, Hildebrand W, Cavett J, Iglehart B, Zachary A (2004). "HLA alleles and haplotypes among the Lakota Sioux: report of the ASHI minority workshops, part III.". Hum Immunol 65 (1): 78–89. doi:10.1016/j.humimm.2003.10.001. PMID 14700599.
- ↑ Lázaro A, Moraes M, Marcos C, Moraes J, Fernández-Viña M, Stastny P (1999). "Evolution of HLA-class I compared to HLA-class II polymorphism in Terena, a South-American Indian tribe.". Hum Immunol 60 (11): 1138–49. doi:10.1016/S0198-8859(99)00092-0. PMID 10600013.
- ↑ "Review Population Study - USA San Antonio Caucasians". Allele frequencies in worldwide population. Retrieved 2008-08-17.
- ↑ Matsuzaka Y, Makino S, Nakajima K, Tomizawa M, Oka A, Bahram S, Kulski J, Tamiya G, Inoko H (2001). "New polymorphic microsatellite markers in the human MHC class III region.". Tissue Antigens 57 (5): 397–404. doi:10.1034/j.1399-0039.2001.057005397.x. PMID 11556964.
- ↑ Kuffner T, Whitworth W, Jairam M, McNicholl J (2003). "HLA class II and TNF genes in African Americans from the Southeastern United States: regional differences in allele frequencies.". Hum Immunol 64 (6): 639–47. doi:10.1016/S0198-8859(03)00056-9. PMID 12770797.
- ↑ 16.0 16.1 Nelson J, Boyer G, Templin D, Lanier A, Barrington R, Nisperos B, Smith A, Mickelson E, Hansen J (1992). "HLA antigens in Tlingit Indians with rheumatoid arthritis.". Tissue Antigens 40 (2): 57–63. doi:10.1111/j.1399-0039.1992.tb01960.x. PMID 1412417.
- ↑ Leffell M, Fallin M, Erlich H, Fernandez-Vĩna M, Hildebrand W, Mack S, Zachary A (2002). "HLA antigens, alleles and haplotypes among the Yup'ik Alaska natives: report of the ASHI Minority Workshops, Part II.". Hum Immunol 63 (7): 614–25. doi:10.1016/S0198-8859(02)00415-9. PMID 12072196.
- ↑ 19.0 19.1 Welinder L, Graugaard B, Madsen M (2000). "HLA antigen and gene frequencies in Eskimos of East Greenland.". Eur J Immunogenet 27 (2): 93–7. doi:10.1046/j.1365-2370.2000.00209.x. PMID 10792425.
- ↑ 20.0 20.1 Lou H, Li H, Kuwayama M, Yashiki S, Fujiyoshi T, Suehara M, Osame M, Yamashita M, Hayami M, Gurtsevich V, Ballas M, Imanishi T, Sonoda S (1998). "HLA class I and class II of the Nivkhi, an indigenous population carrying HTLV-I in Sakhalin, Far Eastern Russia.". Tissue Antigens 52 (5): 444–51. doi:10.1111/j.1399-0039.1998.tb03071.x. PMID 9864034.
- ↑ Sidney J, del Guercio MF, Southwood S, and Sette A. (2002). "The HLA molecules DQA1*0501/B1*0201 and DQA1*0301/B1*0302 share an extensive overlap in peptide binding specificity.". Journal of Immunology 169 (9): 5098–5108. doi:10.4049/jimmunol.169.9.5098. PMID 12391226.
- ↑ Two references:1. "Review Population Study - Iran Yazd Zoroastrians". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Farjadian S, Moqadam F, Ghaderi A (2006). "HLA class II gene polymorphism in Parsees and Zoroastrians of Iran.". Int J Immunogenet 33 (3): 185–91. doi:10.1111/j.1744-313X.2006.00594.x. PMID 16712649.
- ↑ Two references:1. "Review Population Study - Mongolia Khoton Tarialan". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Machulla H, Batnasan D, Steinborn F, Uyar F, Saruhan-Direskeneli G, Oguz F, Carin M, Dorak M (2003). "Genetic affinities among Mongol ethnic groups and their relationship to Turks.". Tissue Antigens 61 (4): 292–9. doi:10.1034/j.1399-0039.2003.00043.x. PMID 12753667.
- ↑ Two references:1. "Review Population Study - Papua New Guinea Madang". Allele frequencies in the worldwide population. Retrieved 2008-08-17.
- ↑ 25.0 25.1 Two references:1. "Review Population Study - China Yunnan Province Yao". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Liu Y, Liu Z, Fu Y, Jia Z, Chen S, Xu A (2006). "Polymorphism of HLA class II genes in Miao and Yao nationalities of Southwest China.". Tissue Antigens 67 (2): 157–9. doi:10.1111/j.1399-0039.2006.00510.x. PMID 16441488.
- ↑ Bannai M, Tokunaga K, Imanishi T, Harihara S, Fujisawa K, Juji T, Omoto K (1996). "HLA class II alleles in Ainu living in Hidaka District, Hokkaidō, northern Japan.". Am J Phys Anthropol 101 (1): 1–9. doi:10.1002/(SICI)1096-8644(199609)101:1<1::AID-AJPA1>3.0.CO;2-Z. PMID 8876810.
- ↑ Two references:1. "Review Population Study - China Shandong Han". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Zhou L, Lin B, Xie Y, Liu Z, Yan W, Xu A (2005). "Polymorphism of human leukocyte antigen-DRB1, -DQB1, and -DPB1 genes of Shandong Han population in China.". Tissue Antigens 66 (1): 37–43. doi:10.1111/j.1399-0039.2005.00418.x. PMID 15982255.
- ↑ Four references:1.Machulla H, Batnasan D, Steinborn F, Uyar F, Saruhan-Direskeneli G, Oguz F, Carin M, Dorak M (2003). "Genetic affinities among Mongol ethnic groups and their relationship to Turks.". Tissue Antigens 61 (4): 292–9. doi:10.1034/j.1399-0039.2003.00043.x. PMID 12753667., 2.Chimge N, Tanaka H, Kashiwase K, Ayush D, Tokunaga K, Saji H, Akaza T, Batsuuri J, Juji T (1997). "The HLA system in the population of Mongolia.". Tissue Antigens 49 (5): 477–83. doi:10.1111/j.1399-0039.1997.tb02782.x. PMID 9174140., 3."Review Population Study - Mongolia Khalkha". Allele frequencies in the worldwide population. Retrieved 2008-08-17. , and 4."Review Population Study - Mongolia Khalkh Ulaanbaatar". Allele frequencies in the worldwide population. Retrieved 2008-08-17.
- ↑ "Review Population Study - Thailand". Allele frequencies in the worldwide population. Retrieved 2008-08-17.
- ↑ Two references:1. "Review Population Study - Russia Siberia Ket Lower Yenisey". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Uinuk-Ool T, Takezaki N, Derbeneva O, Volodko N, Sukernik R (2004). "Variation of HLA class II genes in the Nganasan and Ket, two aboriginal Siberian populations.". Eur J Immunogenet 31 (1): 43–51. doi:10.1111/j.1365-2370.2004.00443.x. PMID 15009181.
- ↑ Lee K, Oh D, Lee C, Yang S (2005). "Allelic and haplotypic diversity of HLA-A, -B, -C, -DRB1, and -DQB1 genes in the Korean population.". Tissue Antigens 65 (5): 437–47. doi:10.1111/j.1399-0039.2005.00386.x. PMID 15853898.
- ↑ Saito S, Ota S, Yamada E, Inoko H, Ota M (2000). "Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population.". Tissue Antigens 56 (6): 522–9. doi:10.1034/j.1399-0039.2000.560606.x. PMID 11169242.
- ↑ Two references, 1."Review Population Study - Russia Siberia Nganasan Dudinka". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Uinuk-Ool T, Takezaki N, Derbeneva O, Volodko N, Sukernik R (2004). "Variation of HLA class II genes in the Nganasan and Ket, two aboriginal Siberian populations.". Eur J Immunogenet 31 (1): 43–51. doi:10.1111/j.1365-2370.2004.00443.x. PMID 15009181.
- ↑ Two references, 1."Review Population Study - China Urumqi Uygur". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Mizuki N, Ohno S, Ando H, Sato T, Imanishi T, Gojobori T, Ishihara M, Goto K, Ota M, Geng Z, Geng L, Li G, Inoko H (1998). "Major histocompatibility complex class II alleles in an Uygur population in the Silk Route of Northwest China.". Tissue Antigens 51 (3): 287–92. doi:10.1111/j.1399-0039.1998.tb03104.x. PMID 9550330.
- ↑ Mizuki M, Ohno S, Ando H, Sato T, Imanishi T, Gojobori T, Ishihara M, Ota M, Geng Z, Geng L, Li G, Kimura M, Inoko H (1997). "Major histocompatibility complex class II alleles in Kazak and Han populations in the Silk Route of northwestern China.". Tissue Antigens 50 (5): 527–34. doi:10.1111/j.1399-0039.1997.tb02909.x. PMID 9389328.
- ↑ Two references, 1."Review Population Study - Russia Siberia Negidal". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Uinuk-Ool T, Takezaki N, Sukernik R, Nagl S, Klein J (2002). "Origin and affinities of indigenous Siberian populations as revealed by HLA class II gene frequencies.". Hum Genet 110 (3): 209–26. doi:10.1007/s00439-001-0668-0. PMID 11935333.
- ↑ Velickovic Z, Delahunt B, Carter J (2002). "HLA-DRB1 and HLA-DQB1 polymorphisms in Pacific Islands populations.". Tissue Antigens 59 (5): 397–406. doi:10.1034/j.1399-0039.2002.590506.x. PMID 12144623.
- ↑ 38.0 38.1 Two references:1. "Review Population Study - Russia Nenets". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2. Evseeva I, Spurkland A, Thorsby E, Smerdel A, Tranebjaerg L, Boldyreva M, Groudakova E, Gouskova I, Alexeev L (2002). "HLA profile of three ethnic groups living in the North-Western region of Russia.". Tissue Antigens 59 (1): 38–43. doi:10.1034/j.1399-0039.2002.590107.x. PMID 11972877.
- ↑ Two references:1. "Review Population Study - Israel%20Gaza%20Palestinians". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Arnaiz-Villena A, Elaiwa N, Silvera C, Rostom A, Moscoso J, Gómez-Casado E, Allende L, Varela P, Martínez-Laso J (2001). "The origin of Palestinians and their genetic relatedness with other Mediterranean populations.". Hum Immunol 62 (9): 889–900. doi:10.1016/S0198-8859(01)00288-9. PMID 11543891. (Retracted. If this is intentional, please replace
{{Retracted}}with{{Retracted|intentional=yes}}.) - ↑ Two references:1. "Review Population Study - Russia Arkhangelsk Pomors". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Evseeva I, Spurkland A, Thorsby E, Smerdel A, Tranebjaerg L, Boldyreva M, Groudakova E, Gouskova I, Alexeev L (2002). "HLA profile of three ethnic groups living in the North-Western region of Russia.". Tissue Antigens 59 (1): 38–43. doi:10.1034/j.1399-0039.2002.590107.x. PMID 11972877.
- ↑ 41.0 41.1 41.2 41.3 Two references:1. "Review Population Study - Sweden". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2. Kimiyoshi, Tsuji; Aizawa M and Sasazuki T (1992). Proceedings of the Eleventh International Histocompatibility Workshop and Conference Held in Yokohoma, Japan, 6–13 November 1991. Oxford: Oxford University Press. ISBN 0-19-262390-7.
- ↑ Bugawan T, Klitz W, Blair A, Erlich H (2000). "High-resolution HLA class I typing in the CEPH families: analysis of linkage disequilibrium among HLA loci". Tissue Antigens 56 (5): 392–404. doi:10.1034/j.1399-0039.2000.560502.x. PMID 11144287.
- ↑ Schipper R, Schreuder G, D'Amaro J, Oudshoorn M (1996). "HLA gene and haplotype frequencies in Dutch blood donors". Tissue Antigens 48 (5): 562–74. doi:10.1111/j.1399-0039.1996.tb02670.x. PMID 8988539.
- ↑ Two references:1. "Review Population Study - Russia Northwest Slavic". Allele frequencies in the worldwide population. Retrieved 2008-08-17. and 2.Kapustin S, Lyshchov A, Alexandrova J, Imyanitov E, Blinov M (1999). "HLA class II molecular polymorphisms in healthy Slavic individuals from North-Western Russia". Tissue Antigens 54 (5): 517–20. doi:10.1034/j.1399-0039.1999.540509.x. PMID 10599891.
- ↑ 45.0 45.1 45.2 45.3 45.4 45.5 45.6 Kimiyoshi, Tsuji; Aizawa M and Sasazuki T (1992). Proceedings of the Eleventh International Histocompatibility Workshop and Conference Held in Yokohoma, Japan, 6–13 November 1991. Oxford: Oxford University Press. ISBN 0-19-262390-7.
- ↑ Evseeva I, Spurkland A, Thorsby E, Smerdel A, Tranebjaerg L, Boldyreva M, Groudakova E, Gouskova I, Alexeev L (2002). "HLA profile of three ethnic groups living in the North-Western region of Russia". Tissue Antigens 59 (1): 38–43. doi:10.1034/j.1399-0039.2002.590107.x. PMID 11972877.
- ↑ Sanchez-Velasco P, Gomez-Casado E, Martinez-Laso J, Moscoso J, Zamora J, Lowy E, Silvera C, Cemborain A, Leyva-Cobián F, Arnaiz-Villena A (2003). "HLA alleles in isolated populations from North Spain: origin of the Basques and the ancient Iberians". Tissue Antigens 61 (5): 384–92. doi:10.1034/j.1399-0039.2003.00041.x. PMID 12753657.
- ↑ Valluri V, Valluei V, Mustafa M, Santhosh A, Middleton D, Alvares M, Alvales M, El Haj E, Gumama O, Abdel-Wareth L, Abdel-Waieth L (2005). "Frequencies of HLA-A, HLA-B, HLA-DR, and HLA-DQ phenotypes in the United Arab Emirates population". Tissue Antigens 66 (2): 107–13. doi:10.1111/j.1399-0039.2005.00441.x. PMID 16029430.
- ↑ Sánchez-Velasco P, Karadsheh N, García-Martín A, Ruíz de Alegría C, Leyva-Cobián F (2001). "Molecular analysis of HLA allelic frequencies and haplotypes in Jordanians and comparison with other related populations". Hum Immunol 62 (9): 901–9. doi:10.1016/S0198-8859(01)00289-0. PMID 11543892.
- ↑ 50.0 50.1 Fort M, de Stefano G, Cambon-Thomsen A, Giraldo-Alvarez P, Dugoujon J, Ohayon E, Scano G, Abbal M (1998). "HLA class II allele and haplotype frequencies in Ethiopian Amhara and Oromo populations". Tissue Antigens 51 (4 Pt 1): 327–36. doi:10.1111/j.1399-0039.1998.tb02971.x. PMID 9583804.
- ↑ Gómez-Casado E, del Moral P, Martínez-Laso J, García-Gómez A, Allende L, Silvera-Redondo C, Longas J, González-Hevilla M, Kandil M, Zamora J, Arnaiz-Villena A (2000). "HLA genes in Arabic-speaking Moroccans: close relatedness to Berbers and Iberians". Tissue Antigens 55 (3): 239–49. doi:10.1034/j.1399-0039.2000.550307.x. PMID 10777099.
- ↑ Ayed K, Ayed-Jendoubi S, Sfar I, Labonne M, Gebuhrer L (2004). "HLA class-I and HLA class-II phenotypic, gene and haplotypic frequencies in Tunisians by using molecular typing data". Tissue Antigens 64 (4): 520–32. doi:10.1111/j.1399-0039.2004.00313.x. PMID 15361135.
- ↑ 53.0 53.1 Renquin J, Sanchez-Mazas A, Halle L, Rivalland S, Jaeger G, Mbayo K, Bianchi F, Kaplan C (2001). "HLA class II polymorphism in Aka Pygmies and Bantu Congolese and a reassessment of HLA-DRB1 African diversity". Tissue Antigens 58 (4): 211–22. doi:10.1034/j.1399-0039.2001.580401.x. PMID 11782272.
- ↑ Pimtanothai N, Hurley C, Leke R, Klitz W, Johnson A (2001). "HLA-DR and -DQ polymorphism in Cameroon". Tissue Antigens 58 (1): 1–8. doi:10.1034/j.1399-0039.2001.580101.x. PMID 11580849.
- ↑ Schnittger L, May J, Loeliger C, Gallin M, Erttmann K, Bienzle U, Kremsner P, Meyer C (1997). "HLA DRB1-DQA1-DQB1 haplotype diversity in two African populations". Tissue Antigens 50 (5): 546–51. doi:10.1111/j.1399-0039.1997.tb02911.x. PMID 9389330.
- ↑ de Pablo R, García-Pacheco J, Vilches C, Moreno M, Sanz L, Rementería M, Puente S, Kreisler M (1997). "HLA class I and class II allele distribution in the Bubi population from the island of Bioko
(Equatorial Guinea)". Tissue Antigens 50 (6): 593–601. doi:10.1111/j.1399-0039.1997.tb02917.x. PMID 9458112. - ↑ Park Y, Tait B, Kawasaki E, Rowley M, Mackay I (2004). "Closer association of IA-2 humoral autoreactivity with HLA DR3/4 than DQB1*0201/*0302 in Korean T1D patients". Annals of the New York Academy of Sciences 1037: 104–9. doi:10.1196/annals.1337.015. PMID 15699500.
- ↑ Hermann R, Turpeinen H, Laine A, Veijola R, Knip M, Simell O, Sipilä I, Akerblom H, Ilonen J (2003). "HLA DR-DQ-encoded genetic determinants of childhood-onset type 1 diabetes in Finland: an analysis of 622 nuclear families". Tissue Antigens 62 (2): 162–9. doi:10.1034/j.1399-0039.2003.00071.x. PMID 12889996.
External links
Coeliac Disease
- Coeliac UK (charity)
- The Celiac Disease Foundation (U.S.)
- The Celiac Sprue Association (U.S.)
- National Digestive Diseases Clearinghouse - page on coeliac disease
- National Foundation for Celiac Awareness (U.S.)
- University of Maryland Center for Celiac Research
Type 1 Diabetes
| ||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||