Glycerol dehydrogenase
| glycerol dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Glycerol dehydrogenase from B. stearothermophilus with glycerol (spheres) PDB 1jqa | |||||||||
| Identifiers | |||||||||
| EC number | 1.1.1.6 | ||||||||
| CAS number | 9028-14-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
Glycerol dehydrogenase (EC 1.1.1.6, also known as NAD+-linked glycerol dehydrogenase, glycerol: NAD+ 2-oxidoreductase, GDH, GlDH, GlyDH) is an enzyme in the oxidoreductase family that utilizes the NAD+ to catalyze the oxidation of glycerol to form glycerone (dihydroxyacetone).[1] [2]

This enzyme is an oxidoreductase, specifically a metal-dependent alcohol dehydrogenase that plays a role in anaerobic glycerol metabolism and has been isolated from a number of bacteria, including Enterobacter aerogenes,[3] Klebsiella aerogenes,[4] Streptococcus faecalis,[5] Erwinia aeroidea,[6]Bacillus megaterium,[7] and Bacillus stearothermophilus. However, most studies of glycerol dehydrogenase have been performed in Bacillus stearothermophilus, (B. stearothermophilus) due to its thermostability and the following structural and functional information will, therefore, refer primarily to the characterization of the enzyme in this bacterium.[8]
Structure
Glycerol dehydrogenase is a homooctamer composed of eight identical monomer subunits made up of a single polypeptide chain of 370 amino acids (molecular weight 42,000 Da). Each subunit contains 9 beta sheets and 14 alpha helices within two distinct domains (N-terminal, residues 1-162 and C-terminal, residues 163-370). The deep cleft formed between these two domains serves as the enzyme’s active site. This active site consists of one bound metal ion, one NAD+ nicotinamide ring binding site, and a substrate binding site.
Research into the structure of B. stearothermophilus shows that the active site contains a divalent cation—zinc ion, Zn2+. This zinc ion forms tetrahedral dipole interactions between the amino acid residues Asp173, His256, and His274 as well as a water molecule.
The NAD+ binding site, resembling the Rossmann fold within the N-terminal domain, extends from the surface of the enzyme to the cleft containing the active site. The nicotinamide ring (the active region of NAD+) binds in a pocket of the cleft consisting of the residues Asp100, Asp123, Ala124, Ser127, Leu129, Val131, Asp173, His174, and Phe247.
Finally, the substrate binding site consists of the residues Asp123, His256, His274 as well as a water molecule.[9]
Function
Encoded by the gene gldA, the enzyme glycerol dehydrogenase, GlyDH catalyzes the oxidation of glycerol to glycerone. Unlike more common pathways utilizing glycerol, GlyDH effectively oxidizes glycerol in anaerobic metabolic pathways under ATP-independent conditions (a useful mechanism in the breakdown of glycerol in bacteria). In addition, GlyDH selectively oxidizes the C2 hydroxyl group to form a ketone rather than a terminal hydroxyl group to form an aldehyde.[10]
Mechanism
While the precise mechanism of this specific enzyme has not yet been characterized, kinetic studies support that GlyDH catalysis of the chemical reaction
- glycerol + NAD+
 glycerone + NADH + H+
glycerone + NADH + H+
is comparable to those of other alcohol dehydrogenases. Therefore, the following mechanism offers a reasonable representation of glycerol oxidation by NAD+.
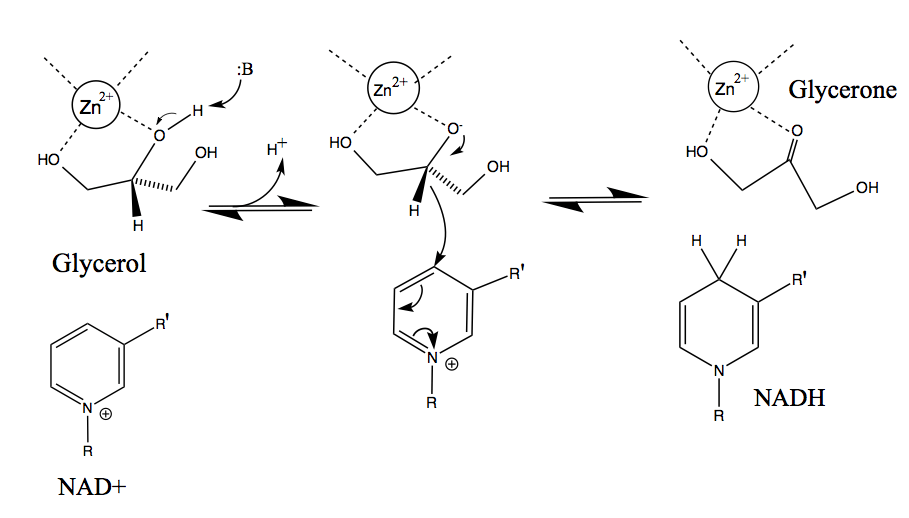
After NAD+ is bound to the enzyme, glycerol substrate binds to the active site in such a way as to have two coordinated interactions between two adjacent hydroxyl groups and the neighboring zinc ion. GlyDH then catalyzes the base-assisted deprotonation of the C2 hydroxyl group, forming an alkoxide. The zinc atom further serves to stabilize the negative charge on the alkoxide intermediate before the excess electron density around the charged oxygen atom shifts to form a double bond with the C2 carbon atom. Hydride is subsequently removed from the secondary carbon and acts as a nucleophile in electron transfer to the NAD+ nicotinamide ring. As a result, the H+ removed by the base is released as a proton into the surrounding solution; followed by the release of the product glycerone, then NADH by GlyDH.[11]
Industrial Implications
As a result of increasing biodiesel production, formation of the byproduct, crude glycerol, has also increased. While glycerol is commonly used in food, pharmaceuticals, cosmetics, and other industries, increased production of crude glycerol has become very expensive to purify and utilize in these industries. Because of this, researchers are interested in finding new economical ways to utilize low-grade glycerol products. Biotechnology is one such technique: using particular enzymes to break down crude glycerol to form products such as 1,3-propanediol, 1,2-propanediol, succinic acid, dihydroxyacetone (glycerone), hydrogen, polyglycerols, and polyesters. As a catalyst for the conversion of glycerol to glycerone, glycerol dehydrogenase is one such enzyme being investigated for this industrial purpose.[12]
A full list of structures solved and deposited in the PDB for this class of enzyme can be found in the info box.
See also
References
- Notes
- ↑ Mallinder, Phillip R.; Pritchard, Andrew; Moir, Anne (1992). "Cloning and characterization of a gene from Bacillus stearothermophilus var. non-diastaticus encoding a glycerol dehydrogenase". Gene 110 (1): 9–16. doi:10.1016/0378-1119(92)90438-U. ISSN 0378-1119. PMID 1339360.
- ↑ Marshall, J. H.; May, J. W.; Sloan, J. (1985). "Purification and Properties of Glycerol: NAD+ 2-Oxidoreductase (Glycerol Dehydrogenase) from Schizosaccharomyces pombe". Microbiology 131 (7): 1581–1588. doi:10.1099/00221287-131-7-1581. ISSN 1350-0872.
- ↑ Burton, Robert Main; Kaplan, Nathan O. (1953). "A DPN SPECIFIC GLYCEROL DEHYDROGENASE FROM AEROBACTER AEROGENES1". Journal of the American Chemical Society 75 (4): 1005–1006. doi:10.1021/ja01100a520. ISSN 0002-7863.
- ↑ Lin ECC and Magasanik B (1960). "The activation of glycerol dehydrogenase from Aerobacter aerogenes by monovalent cations". J. Biol. Chem. 235: 1820–1823. PMID 14417009.
- ↑ Jacobs, NJ and VanDemark PJ. (April 1960). "Comparison of the mechanism of Glycerol Oxidation in Aerobically and Anaerobically Grown Streptococcus Faecalis". Journal of Bacteriology 79 (4): 532–538. PMC 278726. PMID 14406375.
- ↑ Sugiura, Mamoru; Oikawa, Tsutomu; Hirano, Kazuyuki; Shimizu, Hiroshi; Hirata, Fumio (1978). "Purification and some properties of glycerol dehydrogenase from Erwinia aroideae". Chemical & Pharmaceutical Bulletin 26 (3): 716–721. doi:10.1248/cpb.26.716. ISSN 0009-2363.
- ↑ SCHARSCHMIDT, Margrit; PFLEIDERER, Gerhard; METZ, Harald; BRÜMMER, Wolfgang (1983). "Isolierung und Charakterisierung von Glycerin-Dehydrogenase ausBacillus megaterium". Hoppe-Seyler´s Zeitschrift für physiologische Chemie 364 (2): 911–922. doi:10.1515/bchm2.1983.364.2.911. ISSN 0018-4888.
- ↑ Spencer, P.; Bown, K.J.; Scawen, M.D.; Atkinson, T.; Gore, M.G. (1989). "Isolation and characterisation of the glycerol dehydrogenase from Bacillus stearothermophilus". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology 994 (3): 270–279. doi:10.1016/0167-4838(89)90304-X. ISSN 0167-4838.
- ↑ Ruzheinikov, Sergey N.; Burke, Jacky; Sedelnikova, Sveta; Baker, Patrick J.; Taylor, Robert; Bullough, Per A.; Muir, Nicola M.; Gore, Michael G.; Rice, David W. (2001). "Glycerol Dehydrogenase". Structure 9 (9): 789–802. doi:10.1016/S0969-2126(01)00645-1. ISSN 0969-2126. PMID 11566129.
- ↑ Leichus, Betty N.; Blanchard, John S. (1994). "Isotopic Analysis of the Reaction Catalyzed by Glycerol Dehydrogenase". Biochemistry 33 (48): 14642–14649. doi:10.1021/bi00252a033. ISSN 0006-2960. PMID 7981227.
- ↑ Hammes-Schiffer, Sharon; Benkovic, Stephen J. (2006). "Relating Protein Motion to Catalysis". Annual Review of Biochemistry 75 (1): 519–541. doi:10.1146/annurev.biochem.75.103004.142800. ISSN 0066-4154. PMID 16756501.
- ↑ Pachauri, Naresh and He, Brian. (July 2006). "Value-added Utilization of Crude Glycerol from Biodiesel Production: A Survey of Current Research Activities". American Society of Agricultural and Biological Engineers.
- Bibliography
- Asnis RE, Brodie AF (1953). "A glycerol dehydrogenase from Escherichia coli". J. Biol. Chem. 203 (1): 153–9. PMID 13069498.
- Burton RM and Kaplan NO (1953). "A DPN specific glycerol dehydrogenase from Aerobacter aerogenes". J. Am. Chem. Soc. 75 (4): 1005–1007. doi:10.1021/ja01100a520.
- Lin ECC and Magasanik B (1960). "The activation of glycerol dehydrogenase from Aerobacter aerogenes by monovalent cations". J. Biol. Chem. 235: 1820–1823. PMID 14417009.