Glutamyl-tRNA reductase
| glutamyl-tRNA reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.2.1.70 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
A glutamyl-tRNA reductase (EC 1.2.1.70) is an enzyme that catalyzes the chemical reaction
- L-glutamate 1-semialdehyde + NADP+ + tRNAGlu
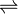 L-glutamyl-tRNAGlu + NADPH + H+
L-glutamyl-tRNAGlu + NADPH + H+
The 3 substrates of this enzyme are L-glutamate 1-semialdehyde, NADP+, and tRNA(Glu), whereas its 3 products are L-glutamyl-tRNA(Glu), NADPH, and H+.
This enzyme belongs to the family of oxidoreductases, to be specific, those acting on the aldehyde or oxo group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is L-glutamate-semialdehyde: NADP+ oxidoreductase (L-glutamyl-tRNAGlu-forming). This enzyme participates in porphyrin and chlorophyll metabolism.
References
- Von Wettstein D, Gough S, Kannangara CG (1995). "Chlorophyll Biosynthesis". Plant. Cell. 7 (7): 1039–1057. doi:10.1105/tpc.7.7.1039. PMC 160907. PMID 12242396.
- Pontoppidan B, Kannangara CG (1994). "Purification and partial characterisation of barley glutamyl-tRNA(Glu) reductase, the enzyme that directs glutamate to chlorophyll biosynthesis". Eur. J. Biochem. 225 (2): 529–37. doi:10.1111/j.1432-1033.1994.00529.x. PMID 7957167.
- Heinz DW, Inokuchi H, Soll D, Jahn D (2002). "Escherichia coli glutamyl-tRNA reductase. Trapping the thioester intermediate". J. Biol. Chem. 277 (50): 48657–63. doi:10.1074/jbc.M206924200. PMID 12370189.