Genetic history of the Iberian Peninsula
The ancestry of modern Iberians (Spanish and Portuguese) is consistent with the geographical situation of the Iberian Peninsula in the south-west corner of Europe. There is a strong prehistoric connection particularly with Atlantic Europe, but also with the wider Mediterranean region, albeit the latter is lesser than regions to the East of the continent (the Balkans) due to Spain being the farthest away from the Bosphorous region considered the main bridge of population expansions into Europe during the Neolithic. On the other hand, Iberia has the strongest genetic proximity in Europe to North Africa, although with strong regional variations. This purportedly results from northward population movements during the seven centuries of Muslim rule within the peninsula. The Basque Region in Northern Spain is genetically distinct as well as typically Atlantic European, holding the least Neolithic/Mediterranean and North African ancestry in Iberia. Modern day Basques are likely to be genetically similar to the Iberian peninsula's paleolithic inhabitants who repopulated Western Europe after the last Ice Age.
Population Genetics: Methods and Limitations
One of the first scholars to perform genetic studies was Luigi Luca Cavalli-Sforza. He used classical genetic markers to analyse DNA by proxy. This method studies differences in the frequencies of particular allelic traits, namely polymorphisms from proteins found within human blood (such as the ABO blood groups, Rhesus blood antigens, HLA loci, immunoglobulins, G-6-P-D isoenzymes, among others). Subsequently his team calculated genetic distance between populations, based on the principle that two populations that share similar frequencies of a trait are more closely related than populations that have more divergent frequencies of the trait.[1]
Since then, population genetics has progressed significantly and studies using direct DNA analysis are now abundant and may use mitochondrial DNA (mtDNA), the non-recombining portion of the Y chromosome (NRY) or autosomal DNA. MtDNA and NRY DNA share some similar features which have made them particularly useful in genetic anthropology. These properties include the direct, unaltered inheritance of mtDNA and NRY DNA from mother to offspring and father to son, respectively, without the 'scrambling' effects of genetic recombination. We also presume that these genetic loci are not affected by natural selection and that the major process responsible for changes in base pairs has been mutation (which can be calculated).[2]
Whereas Y-DNA and mtDNA haplogroups represent but a small component of a person’s DNA pool, autosomal DNA has the advantage of containing hundreds and thousands of examinable genetic loci, thus giving a more complete picture of genetic composition. Descent relationships can only to be determined on a statistical basis, because autosomal DNA undergoes recombination. A single chromosome can record a history for each gene. Autosomal studies are much more reliable for showing the relationships between existing populations but do not offer the possibilities for unraveling their histories in the same way as mtDNA and NRY DNA studies promise, despite their many complications.
Genetic studies operate on numerous assumptions and suffer from methodological limitations such as selection bias and confounding. Phenomenon like genetic drift, foundation and bottleneck effects cause large errors, particularly in haplogroup studies. No matter how accurate the methodology, conclusions derived from such studies are compiled on the basis of how the author envisages their data fits with established archaeological or linguistic theories.
Main genetic compositions
DNA analysis shows that Spanish and Portuguese populations are most closely related to other populations of western Europe.[3][4][5] In a comparison to White Americans of Northwest European descent, Spaniards were found to be more genetically diverse, reflective of the long-standing migratory influences and admixture from other populations found in the Iberian peninsula.[6]
According to Dupanloup et al. (2004) the main components in the European genomes appear to derive from ancestors whose features were similar to those of modern Basques (Paleolithic) and Near Easterners (Neolithic), with average values greater than 35% for both these parental populations, regardless of whether or not molecular information is taken into account. The lowest degree of both Basque and Near Eastern admixture is found in Finland, whereas the highest values are, respectively, 70% ("Basque") in Spain and roughly 60% ("Near Eastern") in the Balkans.[7]
Y-Chromosome DNA
Haplogroup R1b

Y-chromosome analysis[8] had suggested Paleolithic ancestry among populations in the Iberian Peninsula and that Iberia may have played a role in the re-population of western Europe after the last glaciation.[9] This shows an ancestral bond between Iberia and the rest of western Europe, and in particular with Atlantic Europe, which share high frequencies of these haplogroups. R1b1a2, the most common western European haplogroup, arose 4,000 to 8,000 years ago in southwest Asia and later spread to Europe.
Haplogroup composition of the ancient Iberians was very similar to that found in the modern Iberian Peninsula populations, suggesting a long-term genetic continuity since pre-Roman times.[10][11]
Haplogroup E
In human genetics, E-V68, is a major Y chromosome haplogroup found in Africa, Western Asia and Europe, and is in turn part of the larger haplogroup known as E-M35. It is identified by the presence of a single nucleotide polymorphism (SNP) mutation on the Y chromosome known as V68. It is a subject of discussion and study in genetics as well as genetic genealogy, archaeology, and historical linguistics.
E-V68 is dominated by its longer-known subclade E-M78. In various publications, both E-V68 and E-M78 have been referred to by other names, especially phylogenetic names such as "E3b1a" which are designed to show their place on the family tree of all humans. These various names change as new discoveries are made and are discussed below.
Haplogroup I
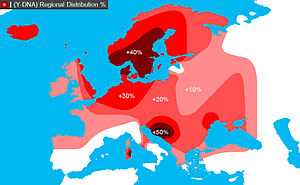
In human genetics, Haplogroup I-M170 is a Y-chromosome DNA haplogroup, a subgroup of haplogroup IJ, itself a derivative of Haplogroup IJK. Y-DNA Haplogroup I-M170 is predominantly a European haplogroup and it is considered as the only native European Haplogroup. Today it represents nearly one-fifth of the population of Europe. It can be found in the majority of present-day European populations with peaks in Northern and South-Eastern Europe. Haplogroup I-M170 Y-chromosomes have also been found among some populations of the Near East, the Caucasus, Northeast Africa and Central Siberia.
Haplogroup R1a

R-M420 (R1a) is a common Y DNA haplogroup in many parts of Eurasia. One sub-clade (branch) of R-M420, R-M17 (R1a1a), is much more common than the others in all major geographical regions. R-M17, defined by the SNP mutation M17, (and sometimes alternatively defined as R-M198), is particularly common in a large region extending from Southern Siberia to Central Europe and Scandinavia.(Underhill 2009)
Mitochondrial DNA
There have been a number of studies about the mitochondrial DNA haplogroups (mtDNA) in Europe. In contrast to Y DNA haplogroups, mtDNA haplogroups did not show as much geographical patterning, but were more evenly ubiquitous. Apart from the outlying Sami, all Europeans are characterized by the predominance of haplogroups H, U and T. The lack of observable geographic structuring of mtDNA may be due to socio-cultural factors, namely the phenomena of polygyny and patrilocality.[12]
The subhaplogroups H1 and H3 have been subject to a more detailed study and would be associated to the Magdalenian expansion from Iberia c. 13,000 years ago:[13]
H1 encompasses an important fraction of Western European mtDNA, reaching its local peak among contemporary Basques (27.8%) and appearing at a high frequency among other Iberians and North Africans. Its frequency is above 10% in many other parts of Europe (France, Sardinia, British Isles, Alps, large portions of Eastern Europe), and above 5% in nearly all the continent. Its subclade H1b is most common in eastern Europe and NW Siberia.[14] So far, the highest frequency of H1 - 61%- has been found among the Tuareg of the Fezzan region in Libya.[15][16]
H3 represents a smaller fraction of European genome than H1 but has a somewhat similar distribution with peak among Basques (13.9%), Galicians (8.3%) and Sardinians (8.5%). Its frequency decreases towards the northeast of the continent, though. Studies have suggested haplogroup H3 is highly protective against AIDS progression.[17]
Autosomal DNA
A 2007 European-wide study including Spanish Basques and Valencian Spaniards, found Iberian populations to cluster the furthest from other continental groups, implying that Iberia holds the most ancient European ancestry. In this study, the most prominent genetic stratification in Europe was found to run from the north to the south-east, while another important axis of differentiation runs east-west across the continent. It also found, despite the differences, that all Europeans are closely related.[18]
North African influence
A number of studies have focused on ascertaining the genetic impact of historical North African population movements into Iberia on the genetic composition of modern Spanish and Portuguese populations. Although initial studies pointed to the Straits of Gibraltar acting more as a genetic barrier than a bridge,[7][19][20] more recent studies point to a substantially higher level of recent North African admixture among Iberians than among other European populations.[21][22][23][24][25][25][26][27]
In terms of autosomal DNA, the most recent study regarding African admixture in Iberian populations was conducted in April 2013 using genome-wide SNP data for over 2000 individuals, concluding that Spain and Portugal hold significantly higher levels of North African than the rest of the European continent. Estimates of shared ancestry averaged between 4 and 20% whereas these did not exceed 2% in other western or southern European populations. However, contrary to past autosomal studies and to what is inferred from Y-Chromosome and Mitochondrial Haplotype frequencies(see below), it does not detect significant levels of Sub-Saharan ancestry in any European population outside the Canary Islands.[28] Indeed, a prior 2011 autosomal study by Moorjani et al. found Sub-Saharan ancestry throughout Europe at ranges of between 1-3%, "the highest proportion of African ancestry in Europe is in Iberia (Portugal 3.2±0.3% and Spain 2.4±0.3%), consistent with inferences based on mitochondrial DNA and Y chromosomes and the observation by Auton et al. that within Europe, the Southwestern Europeans have the highest haplotype-sharing with Africans."[21][25][26]
In terms of paternal Y-Chromosome DNA, recent studies coincide in that Iberia has the greatest presence of the typically Northwest African Y-chromosome haplotype marker E-M81 in Europe.[22][29] as well as Haplotype Va.[30][31] Estimates of Y-Chromosome ancestry vary, with a 2008 study published in the American Journal of Human Genetics using 1140 samples from throughout the Iberian peninsula, giving a proportion of 10.6% North African ancestry.[25][25][26][27] A similar 2009 study of Y-chromosome study with 659 samples from Portugal, 680 from Northern Spain, 37 samples from Andalusia, 915 samples from mainland Italy, and 93 samples from Sicily found significally higher levels of North African male ancestry in Spain, Portugal and Sicily (7.7%, 7.1% and 7.5% respectively) than in Italy (1.7%).[32]
Estimates of Y-Chromosome ancestry vary, with a 2008 study published in the American Journal of Human Genetics using 1140 samples from throughout the Iberian peninsula, stimated that 10.6 percent of modern Iberians (Spain + Portugal) have DNA reflecting some North African ancestors. The mean proportion of identical haplotypes shared between the Sephardic Jewish sample and the Iberian samples is 3.6%, whereas the proportion for those shared between the Moroccan sample and the Iberian samples is 2.8%.[25][25][26][27] A wide-ranging study (published 2007) using 6,501 unrelated Y-chromosome samples from 81 populations found that: "Considering both these E-M78 sub-haplogroups (E-V12, E-V22, E-V65) and the E-M81 haplogroup, the contribution of northern African lineages to the entire male gene pool of Iberia (barring Pasiegos), continental Italy and Sicily can be estimated as 5.6 percent, 3.6 percent and 6.6 percent, respectively".[33] A 2007 study estimated the contribution of northern African lineages to the entire male gene pool of Iberia as 5.6%."[34] As stated, earlier studies (e.g. Bosch et al. 2000) of the Iberian gene-pool had estimated significantly lower levels of North African Ancestry: ".[35]
Recent Mitochondrial DNA studies coincide in that the Iberian Peninsula holds higher levels of typically North African Haplotype U6,[25][26][36][37] as well as higher frequencies of Sub-Saharan African Haplogroup L.[38][13][39][39][40][41] Mean frequency of Haplogroup L lineages reaches 3.83% and the frequency is higher in Portugal (5.83%) than in Spain (2.9%). Furthermore, there is considerable geographic divergence across the peninsula with high frequencies observed for Southern Portugal (10.80%), Central Portugal (9.70%), Western Andalusia (14.6%)[40] and Córdoba (8.30%).[41]
Current debates revolve around whether U6 presence is due to Islamic expansion into the Iberian peninsula or prior population movements[25][25][26][27] and whether Haplogroup L is linked to the slave trade or prior population movements linked to Islamic expansion. A majority of Haplogroup L lineages in Iberia being North African in origin points to the latter.[13][39][42][43][44]
See also
References
- ↑ Cavalli-Sforza (1993, p. 51)
- ↑ Milisauskas (2002, p. 58)
- ↑ Nelis, Mari; Esko, Tõnu; Mägi, Reedik; Zimprich, Fritz; Zimprich, Alexander; Toncheva, Draga; Karachanak, Sena; Piskácková, Tereza; Balascák, Ivan; Peltonen, L; Jakkula, E; Rehnström, K; Lathrop, M; Heath, S; Galan, P; Schreiber, S; Meitinger, T; Pfeufer, A; Wichmann, HE; Melegh, B; Polgár, N; Toniolo, D; Gasparini, P; d'Adamo, P; Klovins, J; Nikitina-Zake, L; Kucinskas, V; Kasnauskiene, J; Lubinski, J; Debniak, T (2009). Fleischer, Robert C., ed. "Genetic Structure of Europeans: A View from the North–East". PLoS ONE 4 (5): e5472. Bibcode:2009PLoSO...4.5472N. doi:10.1371/journal.pone.0005472. PMC 2675054. PMID 19424496.
- ↑ Wade, Nicholas (13 August 2008). "The Genetic Map of Europe". The New York Times. Retrieved 17 October 2009.
- ↑ Novembre, John; Johnson, Toby; Bryc, Katarzyna; Kutalik, Zoltán; Boyko, Adam R.; Auton, Adam; Indap, Amit; King, Karen S.; Bergmann, Sven; Nelson, Matthew R.; Stephens, Matthew; Bustamante, Carlos D. (2008). "Genes mirror geography within Europe". Nature 456 (7218): 98–101. Bibcode:2008Natur.456...98N. doi:10.1038/nature07331. PMC 2735096. PMID 18758442. Lay summary – Gene Expression (31 August 2008).
- ↑ Gayán et al. (2010). "Genetic Structure of the Spanish Population". PMID 20500880.
- ↑ 7.0 7.1 Dupanloup, I.; Bertorelle, G; Chikhi, L; Barbujani, G (2004). "Estimating the Impact of Prehistoric Admixture on the Genome of Europeans". Molecular Biology and Evolution 21 (7): 1361–72. doi:10.1093/molbev/msh135. PMID 15044595.
- ↑ McDonald, J. D. (2005). "Y Haplogroups of the World" (PDF). Retrieved 17 October 2009.
- ↑ Flores, Carlos; Maca-Meyer, Nicole; González, Ana M; Oefner, Peter J; Shen, Peidong; Pérez, Jose A; Rojas, Antonio; Larruga, Jose M; Underhill, Peter A (2004). "Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography". European Journal of Human Genetics 12 (10): 855–63. doi:10.1038/sj.ejhg.5201225. PMID 15280900.
- ↑ Sampietro, M. L.; Caramelli, D.; Lao, O.; Calafell, F.; Comas, D.; Lari, M.; Agustí, B.; Bertranpetit, J.; Lalueza-Fox, C. (2005). "The Genetics of the Pre-Roman Iberian Peninsula: A mtDNA Study of Ancient Iberians". Annals of Human Genetics 69 (Pt 5): 535–48. doi:10.1111/j.1529-8817.2005.00194.x. PMID 16138912.
- ↑ Comas, David; Calafell, Francesc; Benchemsi, Noufissa; Helal, Ahmed; Lefranc, Gerard; Stoneking, Mark; Batzer, Mark A.; Bertranpetit, Jaume; Sajantila, Antti (2000). "Alu insertion polymorphisms in NW Africa and the Iberian Peninsula: evidence for a strong genetic boundary through the Gibraltar Straits". Human Genetics 107 (4): 312–9. doi:10.1007/s004390000370. PMID 11129330.
- ↑ Rosser et al. (2000)
- ↑ 13.0 13.1 13.2 Pereira L, Cunha C, Alves C, Amorim A (April 2005). "African female heritage in Iberia: a reassessment of mtDNA lineage distribution in present times". Human Biology 77 (2): 213–29. doi:10.1353/hub.2005.0041. PMID 16201138.
- ↑ Loogväli EL, Roostalu U, Malyarchuk BA et al. (November 2004). "Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia". Molecular Biology and Evolution 21 (11): 2012–21. doi:10.1093/molbev/msh209. PMID 15254257.
- ↑ Ottoni et al. 2010, "Mitochondrial Haplogroup H1 in North Africa: An Early Holocene Arrival from Iberia", Plosone
- ↑ Ottoni et al., "Table of frequencies of haplogroup H1 in the world", Plosone
- ↑ Hendrickson SL, Hutcheson HB, Ruiz-Pesini E et al. (November 2008). "Mitochondrial DNA Haplogroups influence AIDS Progression". AIDS 22 (18): 2429–39. doi:10.1097/QAD.0b013e32831940bb. PMC 2699618. PMID 19005266.
- ↑ Bauchet, M; McEvoy, B; Pearson, LN; Quillen, EE; Sarkisian, T; Hovhannesyan, K; Deka, R; Bradley, DG; Shriver, MD (2007). "Measuring European Population Stratification with Microarray Genotype Data". The American Journal of Human Genetics 80 (5): 948–56. doi:10.1086/513477. PMC 1852743. PMID 17436249.
- ↑ Bosch, Elena; Calafell, Francesc; Comas, David; Oefner, Peter J.; Underhill, Peter A.; Bertranpetit, Jaume (2001). "High-Resolution Analysis of Human Y-Chromosome Variation Shows a Sharp Discontinuity and Limited Gene Flow between Northwestern Africa and the Iberian Peninsula". The American Journal of Human Genetics 68 (4): 1019–29. doi:10.1086/319521. PMC 1275654. PMID 11254456.
- ↑ Comas, David; Calafell, Francesc; Benchemsi, Noufissa; Helal, Ahmed; Lefranc, Gerard; Stoneking, Mark; Batzer, Mark A.; Bertranpetit, Jaume; Sajantila, Antti (2000). "Alu insertion polymorphisms in NW Africa and the Iberian Peninsula: Evidence for a strong genetic boundary through the Gibraltar Straits". Human Genetics 107 (4): 312–9. doi:10.1007/s004390000370. PMID 11129330.
- ↑ 21.0 21.1 Moorjani P; Patterson N; Hirschhorn JN; Keinan A; Hao L et al. (2011). McVean, Gil, ed. "The History of African Gene Flow into Southern Europeans, Levantines, and Jews". PLoS Genet 7 (4): e1001373. doi:10.1371/journal.pgen.1001373. PMC 3080861. PMID 21533020.
- ↑ 22.0 22.1 Capelli, Cristian; Onofri, Valerio; Brisighelli, Francesca; Boschi, Ilaria; Scarnicci, Francesca; Masullo, Mara; Ferri, Gianmarco; Tofanelli, Sergio; Tagliabracci, Adriano; Gusmao, Leonor; Amorim, Antonio; Gatto, Francesco; Kirin, Mirna; Merlitti, Davide; Brion, Maria; Verea, Alejandro Blanco; Romano, Valentino; Cali, Francesco; Pascali, Vincenzo (2009). "Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe". European Journal of Human Genetics 17 (6): 848–52. doi:10.1038/ejhg.2008.258. PMC 2947089. PMID 19156170.
- ↑ Semino, Ornella; Magri, Chiara; Benuzzi, Giorgia; Lin, Alice A.; Al-Zahery, Nadia; Battaglia, Vincenza; MacCioni, Liliana; Triantaphyllidis, Costas; Shen, Peidong; Oefner, Peter J.; Zhivotovsky, Lev A.; King, Roy; Torroni, Antonio; Cavalli-Sforza, L. Luca; Underhill, Peter A.; Santachiara-Benerecetti, A. Silvana (2004). "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area". The American Journal of Human Genetics 74 (5): 1023–34. doi:10.1086/386295. PMC 1181965. PMID 15069642.
- ↑ Gérard, Nathalie; Berriche, Sala; Aouizérate, Annie; Diéterlen, Florent; Lucotte, Gérard (2006). "North African Berber and Arab Influences in the Western Mediterranean Revealed by Y-Chromosome DNA Haplotypes". Human Biology 78 (3): 307–16. doi:10.1353/hub.2006.0045. PMID 17216803.
- ↑ 25.0 25.1 25.2 25.3 25.4 25.5 25.6 25.7 25.8 25.9 Adams, Susan M.; Bosch, Elena; Balaresque, Patricia L.; Ballereau, Stéphane J.; Lee, Andrew C.; Arroyo, Eduardo; López-Parra, Ana M.; Aler, Mercedes; Grifo, Marina S. Gisbert; Brion, Maria; Carracedo, Angel; Lavinha, João; Martínez-Jarreta, Begoña; Quintana-Murci, Lluis; Picornell, Antònia; Ramon, Misericordia; Skorecki, Karl; Behar, Doron M.; Calafell, Francesc; Jobling, Mark A. (2008). "The Genetic Legacy of Religious Diversity and Intolerance: Paternal Lineages of Christians, Jews, and Muslims in the Iberian Peninsula". The American Journal of Human Genetics 83 (6): 725–36. doi:10.1016/j.ajhg.2008.11.007. PMC 2668061. PMID 19061982. Lay summary – Science News (3 January 2009).
- ↑ 26.0 26.1 26.2 26.3 26.4 26.5 González, Ana M.; Brehm, Antonio; Pérez, José A.; Maca-Meyer, Nicole; Flores, Carlos; Cabrera, Vicente M. (2003). "Mitochondrial DNA affinities at the Atlantic fringe of Europe". American Journal of Physical Anthropology 120 (4): 391–404. doi:10.1002/ajpa.10168. PMID 12627534.
- ↑ 27.0 27.1 27.2 27.3 Giacomo, F.; Luca, F.; Popa, L. O.; Akar, N.; Anagnou, N.; Banyko, J.; Brdicka, R.; Barbujani, G. et al. (2004). "Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe". Human Genetics 115 (5): 357–71. doi:10.1007/s00439-004-1168-9. PMID 15322918.
- ↑ "Gene flow from North Africa contributes to differential human genetic diversity in southern Europe". Proceedings of the National Academy of Sciences of the United States of America 110 (29): 11791–11796. July 16, 2013. doi:10.1073/pnas.1306223110. PMC 3718088. PMID 23733930.
- ↑ Semino, Ornella; Magri, Chiara; Benuzzi, Giorgia; Lin, Alice A.; Al-Zahery, Nadia; Battaglia, Vincenza; MacCioni, Liliana; Triantaphyllidis, Costas; Shen, Peidong; Oefner, Peter J.; Zhivotovsky, Lev A.; King, Roy; Torroni, Antonio; Cavalli-Sforza, L. Luca; Underhill, Peter A.; Santachiara-Benerecetti, A. Silvana (2004). "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area". The American Journal of Human Genetics 74 (5): 1023–34. doi:10.1086/386295. PMC 1181965. PMID 15069642.
- ↑ http://digitalcommons.wayne.edu/humbiol/vol73/iss5/11/
- ↑ Gérard, Nathalie; Berriche, Sala; Aouizérate, Annie; Diéterlen, Florent; Lucotte, Gérard (2006). "North African Berber and Arab Influences in the Western Mediterranean Revealed by Y-Chromosome DNA Haplotypes". Human Biology 78 (3): 307–16. doi:10.1353/hub.2006.0045. PMID 17216803.
- ↑ Capelli, Cristian; Onofri, Valerio; Brisighelli, Francesca; Boschi, Ilaria; Scarnicci, Francesca; Masullo, Mara; Ferri, Gianmarco; Tofanelli, Sergio et al. (2009). "Moors and Saracens in Europe: Estimating the medieval North African male legacy in southern Europe". European Journal of Human Genetics 17 (6): 848–52. doi:10.1038/ejhg.2008.258. PMC 2947089. PMID 19156170. See table
- ↑ Cruciani, F.; La Fratta, R.; Trombetta, B.; Santolamazza, P.; Sellitto, D.; Colomb, E. B.; Dugoujon, J. -M.; Crivellaro, F.; Benincasa, T. (2007). "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12". Molecular Biology and Evolution 24 (6): 1300–1311. doi:10.1093/molbev/msm049. PMID 17351267.
- ↑ Cruciani, F.; La Fratta, R.; Trombetta, B.; Santolamazza, P.; Sellitto, D.; Colomb, E. B.; Dugoujon, J.-M.; Crivellaro, F.; Benincasa, T.; Pascone, R.; Moral, P.; Watson, E.; Melegh, B.; Barbujani, G.; Fuselli, S.; Vona, G.; Zagradisnik, B.; Assum, G.; Brdicka, R.; Kozlov, A. I.; Efremov, G. D.; Coppa, A.; Novelletto, A.; Scozzari, R. (2007). "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12". Molecular Biology and Evolution 24 (6): 1300–11. doi:10.1093/molbev/msm049. PMID 17351267.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1275654/?tool=pubmed
- ↑ Plaza, S.; Calafell, F.; Helal, A.; Bouzerna, N.; Lefranc, G.; Bertranpetit, J.; Comas, D. (2003). "Joining the Pillars of Hercules: mtDNA Sequences Show Multidirectional Gene Flow in the Western Mediterranean". Annals of Human Genetics 67 (Pt 4): 312–28. doi:10.1046/j.1469-1809.2003.00039.x. PMID 12914566.
- ↑ Pereira, Luisa; Cunha, Carla; Alves, Cintia; Amorim, Antonio (2005). "African Female Heritage in Iberia: A Reassessment of mtDNA Lineage Distribution in Present Times". Human Biology 77 (2): 213–29. doi:10.1353/hub.2005.0041. PMID 16201138.
- ↑ http://genome.cshlp.org/content/22/5/821.full#ref-12
- ↑ 39.0 39.1 39.2 Brehm A, Pereira L, Kivisild T, Amorim A (December 2003). "Mitochondrial portraits of the Madeira and Açores archipelagos witness different genetic pools of its settlers". Human Genetics 114 (1): 77–86. doi:10.1007/s00439-003-1024-3. PMID 14513360.
- ↑ 40.0 40.1 http://www.biomedcentral.com/content/pdf/1471-2156-15-11.pdf
- ↑ 41.0 41.1 Pereira, Luisa; Cunha, Carla; Alves, Cintia; Amorim, Antonio (2005). "African Female Heritage in Iberia: A Reassessment of mtDNA Lineage Distribution in Present Times". Human Biology 77 (2): 213–29. doi:10.1353/hub.2005.0041. PMID 16201138.
- ↑ http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1001373
- ↑ Gonzalez et al. (2003)
- ↑ Alvarez et al. (2010). "Mitochondrial DNA Patterns in the Iberian Northern Plateau: Population Dynamics and Substructure of the Zamora Province". PMID 20127843.
| ||||||||||