Force spectroscopy
Force spectroscopy is a set of techniques for the study of the binding forces between individual molecules.[1][2] These methods can be used to measure the mechanical properties of single polymer molecules or proteins, or individual chemical bonds. The name "force spectroscopy", although widely used in the scientific community, is somewhat misleading, because there is no true matter-radiation interaction.[3]
Techniques that can be used to perform force spectroscopy include atomic force microscopy,[2] optical tweezers,[4] magnetic tweezers, microneedles,[5] and biomembranes.[6]
Force spectroscopy measures the behavior of a molecule under stretching or torsional mechanical force. In this way a great deal has been learned in recent years about the mechanochemical coupling in the enzymes responsible for muscle contraction, transport in the cell, energy generation (F1-ATPase), DNA replication and transcription (polymerases), DNA unknotting and unwinding (topoisomerases and helicases).
As a single-molecule technique, as opposed to typical ensemble spectroscopies, it allows a researcher to determine properties of the particular molecule under study. In particular, rare events such as conformational change, which are masked in an ensemble, may be observed.
Experimental techniques
There are many ways to accurately manipulate single molecules. Prominent among these are optical or magnetic tweezers and atomic-force-microscope (AFM) cantilevers. In all of these techniques, a biomolecule, such as protein or DNA, or some other biopolymer has one end bound to a surface and the other to a force sensor. The force sensor is usually a micrometre-sized bead or a cantilever, whose displacement can be measured to determine the force.
Atomic force microscope cantilevers
Molecules adsorbed on a surface are picked up by a microscopic tip (nanometres wide) that is located on the end of an elastic cantilever. In a more sophisticated version of this experiment (Chemical Force Microscopy) the tips are covalently functionalized with the molecules of interest. A piezoelectric controller then pulls up the cantilever. If some force is acting on the elastic cantilever (for example because some molecule is being stretched between the surface and the tip), this will deflect upward (repulsive force) or downward (attractive force). According to Hooke's law, this deflection will be proportional to the force acting on the cantilever. Deflection is measured by the position of a laser beam reflected by the cantilever. This kind of set-up can measure forces as low as 10 pN (10−11 N), and cannot achieve much better resolution only because of thermal noise. The so-called force curve is the graph of force (or more precisely, of cantilever deflection) versus the piezoelectric position on the Z axis. An ideal Hookean spring, for example, would display a straight diagonal force curve. Typically, the force curves observed in the force spectroscopy experiments consist of a contact (diagonal) region where the probe contacts the sample surface, and a non-contact region where the probe is off the sample surface. When the restoring force of the cantilever exceeds tip-sample adhesion force the probe jumps out of contact, and the magnitude of this jump is often used as a measure of adhesion force or rupture force. In general the rupture of a tip-surface bond is a stochastic process; therefore reliable quantification of the adhesion force requires taking multiple individual force curves. The histogram of the adhesion forces obtained in these multiple measurements provides the main data output for force spectroscopy measurement.
Quite often researchers repeat the measurements as a function of the bond loading rate. The resulting graph of the average rupture force as a function of the loading rate is called the force spectrum and forms the basic dataset for the dynamic force spectroscopy. In the ideal case of a single sharp energy barrier for the tip-sample interactions the dynamic force spectrum will show a linear increase of the rupture force as function of a logarithm of the loading rate. The slope of the line is equal to the 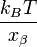 , where
, where 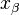 is the distance from the energy minimum to the transition state.
is the distance from the energy minimum to the transition state.
Optical tweezers
Another technique that has been gaining ground for single molecule experiments is the use of optical tweezers for applying mechanical forces on molecules. A strongly focused laser beam has the ability to catch and hold particles (of dielectric material) in a size range from nanometers to micrometers. The trapping action of optical tweezers results from the dipole or optical gradient force on the dielectric sphere. The technique of using a focused laser beam as an atom trap was first applied in 1984 at Bell laboratories. Until then experiments had been carried out using oppositely directed lasers as a means to trap particles. Later experiments, at the same project at Bell laboratories and others since, showed damage-free manipulation on cells using an infrared laser. Thus, the ground was made for biological experiments with optical trapping.
Each technique has its own advantages and disadvantages. For example, AFM cantilevers, can measure angstrom-scale, millisecond events and forces larger than 10 pN. While glass microfibers cannot achieve such fine spatial and temporal resolution, they can measure piconewton forces. Optical tweezers allow the measurement of piconewton forces and nanometer displacements which is an ideal range for many biological experiments. Magnetic tweezers can measure femtonewton forces, and additionally they can also be used to apply torsion.
Applications
Common applications of force spectroscopy are measurements of polymer elasticity, especially biopolymers such as RNA and DNA. Another biophysical application of polymer force spectroscopy is on protein unfolding.[7] Modular proteins can be adsorbed to a gold or (more rarely) mica surface and then stretched. The sequential unfolding of modules is observed as a very characteristic sawtooth pattern of the force vs elongation graph; every tooth corresponds to the unfolding of a single protein module (apart from the last that is generally the detachment of the protein molecule from the tip). Much information about protein elasticity and protein unfolding can be obtained by this technique. Many proteins in the living cell must face mechanical stress.
The other main application of force spectroscopy is the study of mechanical resistance of chemical bonds. In this case, generally the tip is functionalized with a ligand that binds to another molecule bound to the surface. The tip is pushed on the surface, allowing for contact between the two molecules, and then retracted until the newly formed bond breaks up. The force at which the bond breaks up is measured. Since mechanical breaking is a kinetic, stochastic process, the breaking force is not an absolute parameter, but it is a function of both temperature and pulling speed. Low temperatures and high pulling speeds correspond to higher breaking forces. By careful analysis of the breaking force at various pulling speeds, it is possible to map the energy landscape of the chemical bond under mechanical force. This is leading to interesting results in the study of antibody-antigen, protein-protein, protein-living cell interaction and catch bonds.
Recently this technique has been used in cell biology to measure the aggregative stochastic forces created my motor proteins that influence the motion of particles within the cytoplasm. In this, way force spectrum microscopy may be used to better understand the many cellular processes that require the motion of particles within cytoplasm.[8]
References
- ↑ Neuman, Keir C; Nagy, Attila (2008). "Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy". Nature Methods 5 (6): 491–505. doi:10.1038/nmeth.1218. ISSN 1548-7091.
- ↑ 2.0 2.1 Hoffmann, Toni; Dougan, Lorna (2012). "Single molecule force spectroscopy using polyproteins". Chemical Society Reviews 41 (14): 4781. doi:10.1039/c2cs35033e. ISSN 0306-0012.
- ↑ Anna Rita Bizzarri; Salvatore Cannistraro (25 January 2012). Dynamic Force Spectroscopy and Biomolecular Recognition. CRC Press. pp. 1–. ISBN 978-1-4398-6237-7.
- ↑ Jagannathan, B; Marqusee, S (Nov 2013). "Protein folding and unfolding under force". Biopolymers 99: 860–869. doi:10.1002/bip.22321.
- ↑ Kishino, Akiyoshi; Yanagida, Toshio (1988). "Force measurements by micromanipulation of a single actin filament by glass needles". Nature 334 (6177): 74–76. doi:10.1038/334074a0. ISSN 0028-0836.
- ↑ Evans, E.; Ritchie, K.; Merkel, R. (1995). "Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces". Biophysical Journal 68 (6): 2580–2587. doi:10.1016/S0006-3495(95)80441-8. ISSN 0006-3495.
- ↑ Jagannathan, B; Marqusee, S (2012). "Direct observation of a force-induced switch in the anisotropic mechanical unfolding pathway of a protein". PNAS 109: 17820–17825. doi:10.1073/pnas.1201800109.
- ↑ http://www.cell.com/abstract/S0092-8674%2814%2900924-6
Bibliography
- Hugel T, Seitz M. The study of molecular interactions by AFM force spectroscopy. Macromol Rapid Commun 2001; 22:989-1016.
- Janshoff A, Neitzert M, Oberdorfer Y, Fuchs H. Force spectroscopy of molecular systems-single molecule spectroscopy of polymers and biomolecules. Angew Chem Int Ed 2000; 39:3212-3237.
- Oesterhelt, F.; Rief, M.; Gaub, H. E. New J. Phys. 1999; 1:6.1.
- Rief, M. and Grubmueller, H. Force spectroscopy of single biomolecules. Chem. Phys. Chem. 2001; 57:255-261
- Smith, S.; Cui, Y.; Bustamante, C. Science (Washington, D.C.) 1996; 271:795.
- Zhang WK, Zhang X. Single molecule mechanochemistry of macromolecules. Prog Polym Sci 2003; 28:1271-1295.
- Jagannathan, B. and Marqusee, S. Protein Folding and Unfolding under Force. Biopolymers 2013;99:860-869