FOXM1
Forkhead box protein M1 is a protein that in humans is encoded by the FOXM1 gene.[1][2][2] The protein encoded by this gene is a member of the FOX family of transcription factors.[1][3] FOXM1 has been awarded the Molecule of the Year 2010 for its growing potential as a target for cancer diagnosis and therapies.[4]
Function
FOXM1 is known to play a key role in cell cycle progression where endogenous FOXM1 expression peaks at S and G2/M phases.[5] FOXM1-null mouse embryos were neonatal lethal as a result of the development of polyploid cardiomyocytes and hepatocytes, highlighting the role of FOXM1 in mitotic division. More recently a study using transgenic/knockout mouse embryonic fibroblasts and human osteosarcoma cells (U2OS) has shown that FOXM1 regulates expression of a large array of G2/M-specific genes, such as Plk1, cyclin B2, Nek2 and CENPF, and plays an important role in maintenance of chromosomal segregation and genomic stability.[6]
Cancer link
FOXM1 gene is now known as a human proto-oncogene.[7] Abnormal upregulation of FOXM1 is involved in the oncogenesis of basal cell carcinoma, the most common human cancer worldwide.[8] FOXM1 upregulation was subsequently found in the majority of solid human cancers including liver,[9] breast,[10] lung,[11] prostate,[12] cervix of uterus,[13] colon,[14] pancreas,[15] and brain.[16]
Isoforms
There are three FOXM1 isoforms, A, B and C. Isoform FOXM1A has been shown to be a gene transcriptional repressor whereas the remaining isoforms (B and C) are both transcriptional activators. Hence, it is not surprising that FOXM1B and C isoforms have been found to be upregulated in human cancers.[5]
Mechanism of oncogenesis
The exact mechanism of FOXM1 in cancer formation remains unknown. It is thought that upregulation of FOXM1 promotes oncogenesis through abnormal impact on its multiple roles in cell cycle and chromosomal/genomic maintenance. Aberrant upregulation of FOXM1 in primary human skin keratinocytes can directly induce genomic instability in the form of loss of heterozygosity (LOH) and copy number aberrations.[17]
FOXM1 overexpression is involved in early events of carcinogenesis in head and neck squamous cell carcinoma. It has been shown that nicotine exposure directly activates FOXM1 activity in human oral keratinocytes and induced malignant transformation.[18]
Role in stem cell fate
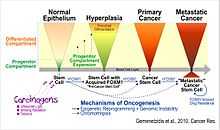
A recent report by the research group which first found that the over-expression of FOXM1 is associated with human cancer, showed that aberrant upregulation of FOXM1 in adult human epithelial stem cells induces a precancer phenotype in a 3D-organotypic tissue regeneration system - a condition similar to human hyperplasia. The authors showed that excessive expression of FOXM1 exploits the inherent self-renewal proliferation potential of stem cells by interfering with the differentiation pathway, thereby expanding the progenitor cell compartment. It was therefore hypothesized that FOXM1 induces cancer initiation through stem/progenitor cell expansion.[19]
Role in epigenome regulations
Given the role in progenitor/stem cells expansion,[19] FOXM1 has been shown to modulate the epigenome. It was found that overexpression of FOXM1 "brain washes" normal cells to adopt cancer-like epigenome. A number of new epigenetic biomarkers influenced by FOXM1 were identified from the study and these were thought to represent epigenetic signature of early cancer development which has potential for early cancer diagnosis and prognosis.[20]
Clinical applications
Precancer initiation and multifaceted oncogenic roles of FOXM1 in a myriad of human cancers render it a highly promising biomarker for cancer diagnostics and anticancer drug development. Hence, FOXM1 gene is currently being exploited for clinical use as biomarker for cancer risk prediction, early cancer screening, molecular diagnostics/prognostics and/or companion diagnostics for personalized therapeutics.
_-_a_FOXM1-Based_Cancer_Detection_Test.jpg)
A number of anti-tumour compounds are being developed to target FOXM1 specifically but none so far has entered clinical trials. Nevertheless, prototype drugs are currently under active research for a number of cancer types.
Interactions
FOXM1 has been shown to interact with CDH1.[22]
See also
References
- ↑ 1.0 1.1 Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG et al. (March 1997). "Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues". Mol. Cell. Biol. 17 (3): 1626–41. PMC 231888. PMID 9032290.
- ↑ 2.0 2.1 Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG et al. (March 1998). "The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization". Genomics 46 (3): 435–42. doi:10.1006/geno.1997.5065. PMID 9441747. Check date values in:
|year= / |date= mismatch(help) - ↑ "Entrez Gene: FOXM1 forkhead box M1".
- ↑ Vincent S. "2010 Molecule of the Year". BioTechniques.
- ↑ 5.0 5.1 Wierstra I, Alves J (December 2007). "FOXM1, a typical proliferation-associated transcription factor". Biol. Chem. 388 (12): 1257–74. doi:10.1515/BC.2007.159. PMID 18020943.
- ↑ Laoukili J, Kooistra MR, Brás A, Kauw J, Kerkhoven RM, Morrison A et al. (February 2005). "FoxM1 is required for execution of the mitotic programme and chromosome stability". Nat. Cell Biol. 7 (2): 126–36. doi:10.1038/ncb1217. PMID 15654331. Vancouver style error (help)
- ↑ Myatt SS, Lam EW (November 2007). "The emerging roles of forkhead box (Fox) proteins in cancer". Nat. Rev. Cancer 7 (11): 847–59. doi:10.1038/nrc2223. PMID 17943136.
- ↑ Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG (15 August 2002). "FOXM1 is a downstream target of Gli1 in basal cell carcinomas". Cancer Res. 62 (16): 4773–80. PMID 12183437.
- ↑ Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM et al. (April 2004). "Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor". Genes Dev. 18 (7): 830–50. doi:10.1101/gad.1200704. PMC 387422. PMID 15082532.
- ↑ Wonsey DR, Follettie MT (June 2005). "Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe". Cancer Res. 65 (12): 5181–9. doi:10.1158/0008-5472.CAN-04-4059. PMID 15958562.
- ↑ Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV et al. (February 2006). "The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer". Cancer Res. 66 (4): 2153–61. doi:10.1158/0008-5472.CAN-05-3003. PMID 16489016.
- ↑ Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV et al. (February 2006). "Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice". Cancer Res. 66 (3): 1712–20. doi:10.1158/0008-5472.CAN-05-3138. PMC 1363687. PMID 16452231.
- ↑ Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN et al. (July 2008). "Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis". J. Pathol. 215 (3): 245–52. doi:10.1002/path.2355. PMID 18464245.
- ↑ Douard R, Moutereau S, Pernet P, Chimingqi M, Allory Y, Manivet P et al. (May 2006). "Sonic Hedgehog-dependent proliferation in a series of patients with colorectal cancer". Surgery 139 (5): 665–70. doi:10.1016/j.surg.2005.10.012. PMID 16701100.
- ↑ Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH (September 2007). "Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells". Cancer Res. 67 (17): 8293–300. doi:10.1158/0008-5472.CAN-07-1265. PMID 17804744.
- ↑ Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF et al. (April 2006). "FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells". Cancer Res. 66 (7): 3593–602. doi:10.1158/0008-5472.CAN-05-2912. PMID 16585184.
- ↑ Teh MT, Gemenetzidis E, Chaplin T, Young BD, Philpott MP (2010). "Upregulation of FOXM1 induces genomic instability in human epidermal keratinocytes". Mol. Cancer 9: 45. doi:10.1186/1476-4598-9-45. PMC 2907729. PMID 20187950.
- ↑ Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M et al. (2009). Jin DY, ed. "FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation". PLoS ONE 4 (3): e4849. doi:10.1371/journal.pone.0004849. PMC 2654098. PMID 19287496.
- ↑ 19.0 19.1 Gemenetzidis E, Elena-Costea D, Parkinson EK, Waseem A, Wan H, Teh MT (2010). "Induction of human epithelial stem/progenitor expansion by FOXM1". Cancer Res. 70 (22): 9515–26. doi:10.1158/0008-5472.CAN-10-2173. PMC 3044465. PMID 21062979.
- ↑ Teh MT, Gemenetzidis E, Patel D, Tariq R, Nadir A, Bahta AW et al. (2012). "FOXM1 induces a global methylation signature that mimics the cancer epigenome in head and neck squamous cell carcinoma". PLoS ONE 7 (3): e34329. doi:10.1371/journal.pone.0034329. PMC 3312909. PMID 22461910.
- ↑ Teh MT, Hutchison IL, Costea DE, Neppelberg E, Liavaag PG, Purdie K et al. (2013). "Exploiting FOXM1-orchestrated molecular network for early squamous cell carcinoma diagnosis and prognosis". Int. J. Cancer 132 (9): 2095–106. doi:10.1002/ijc.27886. PMID 23034676.
- ↑ Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH (Sep 2008). "FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner". Cell Cycle 7 (17): 2720–6. doi:10.4161/cc.7.17.6580. PMID 18758239.
External links
- FOXM1 protein, human at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

