Enzyme catalysis

Enzyme catalysis is the increase in the rate of a chemical reaction by the active site of a protein. The protein catalyst (enzyme) may be part of a multi-subunit complex, and/or may transiently or permanently associate with a Cofactor (e.g. adenosine triphosphate). Catalysis of biochemical reactions in the cell is vital due to the very low reaction rates of the uncatalysed reactions. A key driver of protein evolution is the optimization of such catalytic activities via protein dynamics.[1]
The mechanism of enzyme catalysis is similar in principle to other types of chemical catalysis. By providing an alternative reaction route the enzyme reduces the energy required to reach the highest energy transition state of the reaction. The reduction of activation energy (Ea) increases the amount of reactant molecules that achieve a sufficient level of energy, such that they reach the activation energy and form the product. As with other catalysts, the enzyme is not consumed during the reaction (as a substrate is) but is recycled such that a single enzyme performs many rounds of catalysis.
Induced fit
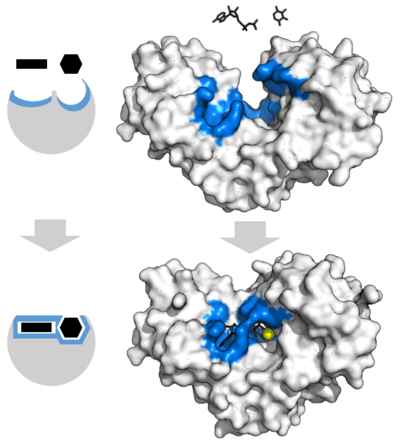

The favored model for the enzyme-substrate interaction is the induced fit model.[2] This model proposes that the initial interaction between enzyme and substrate is relatively weak, but that these weak interactions rapidly induce conformational changes in the enzyme that strengthen binding.
The advantages of the induced fit mechanism arise due to the stabilizing effect of strong enzyme binding. There are two different mechanisms of substrate binding: uniform binding, which has strong substrate binding, and differential binding, which has strong transition state binding. The stabilizing effect of uniform binding increases both substrate and transition state binding affinity, while differential binding increases only transition state binding affinity. Both are used by enzymes and have been evolutionarily chosen to minimize the Ea of the reaction. Enzymes that are saturated, that is, have a high affinity substrate binding, require differential binding to reduce the Ea, whereas small substrate unbound enzymes may use either differential or uniform binding.
These effects have led to most proteins using the differential binding mechanism to reduce the Ea, so most proteins have high affinity of the enzyme to the transition state. Differential binding is carried out by the induced fit mechanism - the substrate first binds weakly, then the enzyme changes conformation increasing the affinity to the transition state and stabilizing it, so reducing the activation energy to reach it.
It is important to clarify, however, that the induced fit concept cannot be used to rationalize catalysis. That is, the chemical catalysis is defined as the reduction of Ea‡ (when the system is already in the ES‡) relative to Ea‡ in the uncatalyzed reaction in water (without the enzyme). The induced fit only suggests that the barrier is lower in the closed form of the enzyme but does not tell us what the reason for the barrier reduction is.
Induced fit may be beneficial to the fidelity of molecular recognition in the presence of competition and noise via the conformational proofreading mechanism .[3]
Mechanisms of an alternative reaction route
These conformational changes also bring catalytic residues in the active site close to the chemical bonds in the substrate that will be altered in the reaction. After binding takes place, one or more mechanisms of catalysis lowers the energy of the reaction's transition state, by providing an alternative chemical pathway for the reaction. There are six possible mechanisms of "over the barrier" catalysis as well as a "through the barrier" mechanism:
Proximity and orientation
This increases the rate of the reaction as enzyme-substrate interactions align reactive chemical groups and hold them close together. This reduces the entropy of the reactants and thus makes reactions such as ligations or addition reactions more favorable, there is a reduction in the overall loss of entropy when two reactants become a single product.
This effect is analogous to an effective increase in concentration of the reagents. The binding of the reagents to the enzyme gives the reaction intramolecular character, which gives a massive rate increase.
| For example: |
| Similar reactions will occur far faster if the reaction is intramolecular. |
 |
| The effective concentration of acetate in the intramolecular reaction can be estimated as k2/k1 = 2 x 105 Molar. |
However, the situation might be more complex, since modern computational studies have established that traditional examples of proximity effects cannot be related directly to enzyme entropic effects.[4][5][6] Also, the original entropic proposal[7] has been found to largely overestimate the contribution of orientation entropy to catalysis.[8]
Proton donors or acceptors
Proton donors and acceptors, i.e. acids and base may donate and accept protons in order to stabilize developing charges in the transition state.This typically has the effect of activating nucleophile and electrophile groups, or stabilizing leaving groups. Histidine is often the residue involved in these acid/base reactions, since it has a pKa close to neutral pH and can therefore both accept and donate protons.
Many reaction mechanisms involving acid/base catalysis assume a substantially altered pKa. This alteration of pKa is possible through the local environment of the residue.
| Conditions | Acids | Bases |
|---|---|---|
| Hydrophobic environment | Increase pKa | Decrease pKa |
| Adjacent residues of like charge | Increase pKa | Decrease pKa |
| Salt bridge (and hydrogen bond) formation |
Decrease pKa | Increase pKa |
pKa can also be influenced significantly by the surrounding environment, to the extent that residues which are basic in solution may act as proton donors, and vice versa.
| For example: |
| Catalytic triad of a serine protease |
 |
| The initial step of the serine protease catalytic mechanism involves the histidine of the active site accepting a proton from the serine residue. This prepares the serine as a nucleophile to attack the amide bond of the substrate. This mechanism includes donation of a proton from serine (a base, pKa 14) to histidine (an acid, pKa 6), made possible due to the local environment of the bases. |
It is important to clarify that the modification of the pKa’s is a pure part of the electrostatic mechanism.[9] Furthermore, the catalytic effect of the above example is mainly associated with the reduction of the pKa of the oxyanion and the increase in the pKa of the histidine, while the proton transfer from the serine to the histidine is not catalyzed significantly, since it is not the rate determining barrier.[10]
Electrostatic catalysis
Stabilization of charged transition states can also be by residues in the active site forming ionic bonds (or partial ionic charge interactions) with the intermediate. These bonds can either come from acidic or basic side chains found on amino acids such as lysine, arginine, aspartic acid or glutamic acid or come from metal cofactors such as zinc. Metal ions are particularly effective and can reduce the pKa of water enough to make it an effective nucleophile.
Systematic computer simulation studies established that electrostatic effects give, by far, the largest contribution to catalysis.[9] In particular, it has been found that enzyme provides an environment which is more polar than water, and that the ionic transition states are stabilized by fixed dipoles. This is very different from transition state stabilization in water, where the water molecules must pay with "reorganization energy".[11] In order to stabilize ionic and charged states. Thus, the catalysis is associated with the fact that the enzyme polar groups are preorganized [12]
The magnitude of the electrostatic field exerted by an enzyme's active site has been shown to be highly correlated with the enzyme's catalytic rate enhancement[13][14]
Binding of substrate usually excludes water from the active site, thereby lowering the local dielectric constant to that of an organic solvent. This strengthens the electrostatic interactions between the charged/polar substrates and the active sites. In addition, studies have shown that the charge distributions about the active sites are arranged so as to stabilize the transition states of the catalyzed reactions. In several enzymes, these charge distributions apparently serve to guide polar substrates toward their binding sites so that the rates of these enzymatic reactions are greater than their apparent diffusion-controlled limits.
| For example: |
| Carboxypeptidase catalytic mechanism |
 |
| The tetrahedral intermediate is stabilised by a partial ionic bond between the Zn2+ ion and the negative charge on the oxygen. |
Covalent catalysis
Covalent catalysis involves the substrate forming a transient covalent bond with residues in the active site or with a cofactor. This adds an additional covalent intermediate to the reaction, and helps to reduce the energy of later transition states of the reaction. The covalent bond must, at a later stage in the reaction, be broken to regenerate the enzyme. This mechanism is utilised by the catalytic triad of enzymes such as proteases like chymotrypsin and trypsin, where an acyl-enzyme intermediate is formed. Schiff base formation using the free amine from a lysine residue is another mechanism, as seen in the enzyme aldolase during glycolysis.
Some enzymes utilize non-amino acid cofactors such as pyridoxal phosphate (PLP) or thiamine pyrophosphate (TPP) to form covalent intermediates with reactant molecules.[15][16] Such covalent intermediates function to reduce the energy of later transition states, similar to how covalent intermediates formed with active site amino acid residues allow stabilization, but the capabilities of cofactors allow enzymes to carryout reactions that amino acid side residues alone could not. Enzymes utilizing such cofactors include the PLP-dependent enzyme aspartate transaminase and the TPP-dependent enzyme pyruvate dehydrogenase.[17][18]
It is important to clarify that covalent catalysis does correspond in most cases to simply the use of a specific mechanism rather than to true catalysis.[9] For example, the energetics of the covalent bond to the serine molecule in chymotrypsin should be compared to the well-understood covalent bond to the nucleophile in the uncatalyzed solution reaction. A true proposal of a covalent catalysis (where the barrier is lower than the corresponding barrier in solution) would require, for example, a partial covalent bond to the transition state by an enzyme group (e.g., a very strong hydrogen bond), and such effects do not contribute significantly to catalysis.
Bond strain
This is the principal effect of induced fit binding, where the affinity of the enzyme to the transition state is greater than to the substrate itself. This induces structural rearrangements which strain substrate bonds into a position closer to the conformation of the transition state, so lowering the energy difference between the substrate and transition state and helping catalyze the reaction.
However, the strain effect is, in fact, a ground state destabilization effect, rather than transition state stabilization effect.[19][9] Furthermore, enzymes are very flexible and they cannot apply large strain effect.[20]
In addition to bond strain in the substrate, bond strain may also be induced within the enzyme itself to activate residues in the active site.
| For example: |
| Substrate, bound substrate, and transition state conformations of lysozyme. |
| |
| The substrate, on binding, is distorted from the half chair conformation of the hexose ring (because of the steric hindrance with amino acids of the protein forcing the equatorial c6 to be in the axial position) into the chair conformation[21] |
Quantum tunneling
These traditional "over the barrier" mechanisms have been challenged in some cases by models and observations of "through the barrier" mechanisms (quantum tunneling). Some enzymes operate with kinetics which are faster than what would be predicted by the classical ΔG‡. In "through the barrier" models, a proton or an electron can tunnel through activation barriers.[22][23] Quantum tunneling for protons has been observed in tryptamine oxidation by aromatic amine dehydrogenase.[24]
Interestingly, quantum tunneling does not appear to provide a major catalytic advantage, since the tunneling contributions are similar in the catalyzed and the uncatalyzed reactions in solution.[23][25][26][27] However, the tunneling contribution (typically enhancing rate constants by a factor of ~1000[24] compared to the rate of reaction for the classical 'over the barrier' route) is likely crucial to the viability of biological organisms. This emphasizes the general importance of tunneling reactions in biology.
In 1971-1972 the first quantum-mechanical model of enzyme catalysis was formulated.[28][29]
Active enzyme
The binding energy of the enzyme-substrate complex cannot be considered as an external energy which is necessary for the substrate activation. The enzyme of high energy content may firstly transfer some specific energetic group X1 from catalytic site of the enzyme to the final place of the first bound reactant, then another group X2 from the second bound reactant (or from the second group of the single reactant) must be transferred to active site to finish substrate conversion to product and enzyme regeneration.[30]
We can present the whole enzymatic reaction as a two coupling reactions:
S1 + EX1 => S1EX1 => P1 + EP2 (1) S2 + EP2 => S2EP2 => P2 + EX2 (2)
It may be seen from reaction (1) that the group X1 of the active enzyme appears in the product due to possibility of the exchange reaction inside enzyme to avoid both electrostatic inhibition and repulsion of atoms. So we represent the active enzyme as a powerful reactant of the enzymatic reaction. The reaction (2) shows incomplete conversion of the substrate because its group X2 remains inside enzyme. This approach as idea had formerly proposed relying on the hypothetical extremely high enzymatic conversions (catalytically perfect enzyme).[31]
The crucial point for the verification of the present approach is that the catalyst must be a complex of the enzyme with the transfer group of the reaction. This chemical aspect is supported by the well-studied mechanisms of the several enzymatic reactions. Let us consider the reaction of peptide bond hydrolysis catalyzed by a pure protein α-chymotrypsin (an enzyme acting without a cofactor), which is a well-studied member of the serine proteases family, see.[32]
We present the experimental results for this reaction as two chemical steps:
S1 + EH => P1 + EP2 (3) EP2 + H–O–H => EH + P2 (4)
where S1 is a polypeptide, P1 and P2 are products. The first chemical step (3) includes the formation of a covalent acyl-enzyme intermediate. The second step (4) is the deacylation step. It is important to note that the group H+, initially found on the enzyme, but not in water, appears in the product before the step of hydrolysis, therefore it may be considered as an additional group of the enzymatic reaction.
Thus, the reaction (3) shows that the enzyme acts as a powerful reactant of the reaction. According to the proposed concept, the H transport from the enzyme promotes the first reactant conversion, breakdown of the first initial chemical bond (between groups P1 and P2). The step of hydrolysis leads to a breakdown of the second chemical bond and regeneration of the enzyme.
The proposed chemical mechanism does not depend on the concentration of the substrates or products in the medium. However, a shift in their concentration mainly causes free energy changes in the first and final steps of the reactions (1) and (2) due to the changes in the free energy content of every molecule, whether S or P, in water solution. This approach is in accordance with the following mechanism of muscle contraction. The final step of ATP hydrolysis in skeletal muscle is the product release caused by the association of myosin heads with actin.[33] The closing of the actin-binding cleft during the association reaction is structurally coupled with the opening of the nucleotide-binding pocket on the myosin active site.[34]
Notably, the final steps of ATP hydrolysis include the fast release of phosphate and the slow release of ADP.[35][36] The release of a phosphate anion from bound ADP anion into water solution may be considered as an exergonic reaction because the phosphate anion has low molecular mass.
Thus, we arrive at the conclusion that the primary release of the inorganic phosphate H2PO4− leads to transformation of a significant part of the free energy of ATP hydrolysis into the kinetic energy of the solvated phosphate, producing active streaming. This assumption of a local mechano-chemical transduction is in accord with Tirosh’s mechanism of muscle contraction, where the muscle force derives from an integrated action of active streaming created by ATP hydrolysis.[37][38]
Examples of catalytic mechanisms
In reality, most enzyme mechanisms involve a combination of several different types of catalysis.
Triose phosphate isomerase
Triose phosphate isomerase (EC 5.3.1.1) catalyses the reversible interconvertion of the two triose phosphates isomers dihydroxyacetone phosphate and D-glyceraldehyde 3-phosphate.
Trypsin
Trypsin (EC 3.4.21.4) is a serine protease that cleaves protein substrates at lysine and arginine amino acid residues using a catalytic triad of active site residues to perform nucleophilic, covalent catalysis.
Aldolase
Aldolase (EC 4.1.2.13) catalyses the breakdown of fructose 1,6-bisphosphate (F-1,6-BP) into glyceraldehyde 3-phosphate and dihydroxyacetone phosphate (DHAP).
Enzyme diffusivity
The advent of single-molecule studies led in the 2010s to the observation that the movement of untethered enzymes increases with increasing substrate concentration and increasing reaction enthalpy.[39] Subsequent observations suggest that this increase in diffusivity is driven by transient displacement of the enzyme's center of mass, resulting in a "recoil effect that propels the enzyme".[40]
Reaction Similarity
Similarity between enzymatic reactions (EC) can be calculated by using bond changes, reaction centres or substructure metrics (EC-BLAST).[41]
See also
- Catalytic triad
- Enzyme assay
- Enzyme kinetics
- Protein dynamics
- Quantum tunnelling
- The Proteolysis Map
- Time resolved crystallography
- Enzyme promiscuity
References
- ↑ Kamerlin, S. C.; Warshel, A (2010). "At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis?". Proteins: Structure, Function, and Bioinformatics 78 (6): 1339–75. doi:10.1002/prot.22654. PMC 2841229. PMID 20099310.
- ↑ Koshland DE (February 1958). "Application of a Theory of Enzyme Specificity to Protein Synthesis". Proc. Natl. Acad. Sci. U.S.A. 44 (2): 98–104. Bibcode:1958PNAS...44...98K. doi:10.1073/pnas.44.2.98. PMC 335371. PMID 16590179.

- ↑ Savir Y; Tlusty T (2007). Scalas, Enrico, ed. "Conformational Proofreading: The Impact of Conformational Changes on the Specificity of Molecular Recognition" (PDF). PLOS ONE 2 (5): e468. Bibcode:2007PLoSO...2..468S. doi:10.1371/journal.pone.0000468. PMC 1868595. PMID 17520027.

- ↑ Stanton, R.V.; Perakyla, M.; Bakowies, D.; Kollman, P.A. (1998). "Combined ab initio and Free Energy Calculations To Study Reactions in Enzymes and Solution: Amide Hydrolysis in Trypsin and Aqueous Solution". J. Am. Chem. Soc 120 (14): 3448–3457. doi:10.1021/ja972723x.
- ↑ Kuhn, B.; Kollman, P.A. (2000). "QM-FE and Molecular Dynamics Calculations on Catechol O-Methyltransferase: Free Energy of Activation in the Enzyme and in Aqueous Solution and Regioselectivity of the Enzyme-Catalyzed Reaction". J. Am. Chem. Soc 122 (11): 2586–2596. doi:10.1021/ja992218v.
- ↑ Bruice, T.C.; Lightstone, F.C. (1999). "Ground State and Transition State Contributions to the Rates of Intramolecular and Enzymatic Reactions". Acc. Chem. Res. 32 (2): 127–136. doi:10.1021/ar960131y.
- ↑ Page, M.I.; Jencks, W.P. (1971). "Entropic Contributions to Rate Accelerations in Enzymic and Intramolecular Reactions and the Chelate Effect". Proc. Natl. Acad. Sci. USA 1971 (68): 1678–1683. Bibcode:1971PNAS...68.1678P. doi:10.1073/pnas.68.8.1678. PMC 389269. PMID 5288752.
- ↑ Warshel, A.; Parson, W.W. (2001). "Dynamics of Biochemical and Biophysical Reactions: Insight from Computer Simulations". Quart. Rev. Biophys 34: 563–679. doi:10.1017/s0033583501003730.
- ↑ 9.0 9.1 9.2 9.3 Warshel A., Sharma P.K., Kato M., Xiang Y., Liu H., and Olsson M.H.M. "Electrostatic Basis of Enzyme Catalysis", Chem. Rev. 2006, 106: 3210-3235.
- ↑ Warshel, A.; Naray-Szabo, G.; Sussman, F.; Hwang, J.-K. (1989). "How do Serine Proteases Really Work?". Biochemistry 1989 (28): 3629–37. doi:10.1021/bi00435a001. PMID 2665806.
- ↑ Marcus R. A. "On the Theory of Electron-Transfer Reactions. VI. Unified Treatment for Homogeneous and Electrode Reactions" J. Chem. Phys. 1965 43:679-701
- ↑ Warshel A. "Energetics of Enzyme Catalysis", Proc. Natl. Acad. Sci. USA, 1978 75: 5250
- ↑ "How Enzymes Work"
- ↑ "EXTREME ELECTRIC FIELDS POWER CATALYSIS IN THE ACTIVE SITE OF KETOSTEROID ISOMERASE",
- ↑ Toney, M. D. "Reaction specificity in pyridoxal enzymes." Archives of biochemistry and biophysics (2005) 433: 279-287
- ↑ Micronutrient Information Center, Oregon State University
- ↑ Voet, Donald; Judith Voet (2004). Biochemistry. John Wiley & Sons Inc. pp. 986–989. ISBN 0-471-25090-2.
- ↑ Voet, Donald; Judith Voet (2004). Biochemistry. John Wiley & Sons Inc. pp. 604–606. ISBN 0-471-25090-2.
- ↑ Jencks, William P. (1987) [1969]. Catalysis in Chemistry and Enzymology. McGraw-Hill series in advanced chemistry (reprint ed.). New York: Dover Publications. ISBN 9780486654607.
- ↑ Warshel, A.; Levitt, M. (1976). "Theoretical Studies of Enzymatic Reactions: Dielectric Electrostatic and Steric Stabilization of the Carbonium Ion in the Reaction of Lysozyme". Journal of Molecular Biology 103 (2): 227–49. doi:10.1016/0022-2836(76)90311-9. PMID 985660.

- ↑ <fundamentals of biochemistry Voet, voet and Pratt 4th edition>, which is similar in shape to the transition state.
- ↑ Garcia-Viloca, M; Gao, J; Karplus, M; Truhlar, DG (2004). "How enzymes work: analysis by modern rate theory and computer simulations". Science 303 (5655): 186–95. Bibcode:2004Sci...303..186G. doi:10.1126/science.1088172. PMID 14716003.
- ↑ 23.0 23.1 Olsson, MH; Siegbahn, PE; Warshel, A (2004). "Simulations of the large kinetic isotope effect and the temperature dependence of the hydrogen atom transfer in lipoxygenase". Journal of the American Chemical Society 126 (9): 2820–8. doi:10.1021/ja037233l. PMID 14995199.
- ↑ 24.0 24.1 Masgrau, L; Roujeinikova, A; Johannissen, LO; Hothi, P; Basran, J; Ranaghan, KE; Mulholland, AJ; Sutcliffe, MJ et al. (2006). "Atomic description of an enzyme reaction dominated by proton tunneling". Science 312 (5771): 237–41. Bibcode:2006Sci...312..237M. doi:10.1126/science.1126002. PMID 16614214.
- ↑ Hwang, J.-K.; Warshel, A. "How important are quantum mechanical nuclear motions in enzyme catalysis". J. Am. Chem. Soc 1996 (118): 11745–11751.
- ↑ Ball, P. (2004). "Enzymes: By chance, or by design?". Nature 431 (7007): 396. Bibcode:2004Natur.431..396B. doi:10.1038/431396a.
- ↑ Olsson, M.H.M.; Parson, W.W.; Warshel, A. (2006). "Dynamical Contributions to Enzyme Catalysis: Critical Tests of A Popular Hypothesis". Chem. Rev. 106 (5): 1737–1756. doi:10.1021/cr040427e. PMID 16683752.
- ↑ Volkenshtein M.V., Dogonadze R.R., Madumarov A.K., Urushadze Z.D., Kharkats Yu.I. Theory of Enzyme Catalysis.- Molekuliarnaya Biologia, Moscow, 6, 1972, 431-439
- ↑ Volkenshtein M.V., Dogonadze R.R., Madumarov A.K., Urushadze Z.D., Kharkats Yu.I. Electronic and Conformational Interactions in Enzyme Catalysis. In: E.L. Andronikashvili (Ed.), Konformatsionnie Izmenenia Biopolimerov v Rastvorakh, Publishing House "Nauka", Moscow, 1973, 153-157
- ↑ Foigel, A.G. (2011). "Is the enzyme a powerful reactant of the biochemical reaction?". Mol. Cell. Biochem 352: 87-89 Foigel, Alexander G. (2011). "Is the enzyme a powerful reactant of the biochemical reaction?". Molecular and Cellular Biochemistry 352 (1–2): 87–9. doi:10.1007/s11010-011-0742-4. PMID 21318350.
- ↑ Fogel, A.G. (1982). "Cooperativity of enzymatic reactions and molecular aspects of energy transduction". Mol. Cell. Biochem 47: 59–64. doi:10.1007/bf00241567.
- ↑ Hengge, AC, Stein, RL. (2004). "Role of protein conformational mobility in enzyme catalysis: acylation of alpha-chymotrypsin by specific peptide substrates". Biochemistry 43 : 742-747
- ↑ Lymn, RW; Taylor, EW. (1971). "Mechanism of adenosine triphosphate hydrolysis by actomyosin". Biochemistry 10: 4617–4624. doi:10.1021/bi00801a004.
- ↑ Holmes, KC; Angert, I; Kull, FG; Jahn, W; Schroder, RR. (2003). "Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide". Nature 425: 423–427. doi:10.1038/nature02005.
- ↑ Siemankowski, RF; Wiseman, MO; White, HD. (1985). "ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle". Proc. Natl. Acad. Sci. USA 82: 658–662. doi:10.1073/pnas.82.3.658.
- ↑ White, HD; Belknap, B; Webb, MR. (1997). "Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate". Biochemistry 36: 11828–11836. doi:10.1021/bi970540h.
- ↑ Tirosh, R; Low, WZ; Oplatka, A. (1990). "Translational motion of actin filaments in the presence of heavy meromyosin and MgATP as measured by Doppler broadening of laser light scattering". Biochim. Biophys. Acta 1037: 274–280. doi:10.1016/0167-4838(90)90025-b.
- ↑ Tirosh, R. (2006). "Ballistic protons and microwave-induced water solutions (solitons) in bioenergetic transformations". Int. J. Mol. Sci. 7: 320–345. doi:10.3390/i7090320.
- ↑ Muddana, Hari S.; Sengupta, Samudra; Mallouk, Thomas E. et al. (28 January 2010). "Substrate Catalysis Enhances Single-Enzyme Diffusion". Journal of the American Chemical Society 132 (7): 2110–1. doi:10.1021/ja908773a.

- ↑ Riedel, Clement; Gabizon, Ronen; Wilson, Christian A. M. et al. (8 January 2015). "The heat released during catalytic turnover enhances the diffusion of an enzyme". Nature 517: 227–30. doi:10.1038/nature14043. Lay summary – Nature: News & Views (8 January 2015).

- ↑ Rahman, SA; Cuesta, SM; Furnham, N; Holliday, GL; Thornton, JM (2014). "EC-BLAST: a tool to automatically search and compare enzyme reactions". Nature Methods 11: 171–174. doi:10.1038/nmeth.2803. PMID 24412978.
Further reading
- Alan Fersht, Structure and Mechanism in Protein Science : A Guide to Enzyme Catalysis and Protein Folding. W. H. Freeman, 1998. ISBN 0-7167-3268-8
- Dedicated issue of Philosophical Transactions B on Quantum catalysis in enzymes freely available.
| ||||||||||||||||||||||
| |||||||||||||||||||||||||