EPSP synthase
| EPSP Synthase (3-phosphoshikimate 1-carboxyvinyltransferase) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
EPSP synthase liganded with shikimate.[1] | |||||||||
| Identifiers | |||||||||
| EC number | 2.5.1.19 | ||||||||
| CAS number | 9068-73-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
| EPSP synthase (3-phosphoshikimate 1-carboxyvinyltransferase) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Ribbon diagram of EPSP synthase | |||||||||
| Identifiers | |||||||||
| Symbol | EPSP_synthase | ||||||||
| Pfam | PF00275 | ||||||||
| InterPro | IPR001986 | ||||||||
| PROSITE | PDOC00097 | ||||||||
| SCOP | 1eps | ||||||||
| SUPERFAMILY | 1eps | ||||||||
| |||||||||
5-enolpyruvylshikimate-3-phosphate (EPSP) synthase is an enzyme that catalyzes the chemical reaction:
- phosphoenolpyruvate + 3-phosphoshikimate
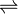 phosphate + 5-enolpyruvylshikimate-3-phosphate (EPSP)
phosphate + 5-enolpyruvylshikimate-3-phosphate (EPSP)
Thus, the two substrates of this enzyme are phosphoenolpyruvate and 3-phospho-shikimate, whereas its two products are phosphate and 5-enolpyruvylshikimate-3-phosphate.
Nomenclature
The enzyme belongs to the family of transferases, to be specific those transferring aryl or alkyl groups other than methyl groups. The systematic name of this enzyme class is phosphoenolpyruvate:3-phosphoshikimate 5-O-(1-carboxyvinyl)-transferase. Other names in common use include:
- 5-enolpyruvylshikimate-3-phosphate synthase,
- 3-enolpyruvylshikimate 5-phosphate synthase,
- 3-enolpyruvylshikimic acid-5-phosphate synthetase,
- 5'-enolpyruvylshikimate-3-phosphate synthase,
- 5-enolpyruvyl-3-phosphoshikimate synthase,
- 5-enolpyruvylshikimate-3-phosphate synthetase,
- 5-enolpyruvylshikimate-3-phosphoric acid synthase,
- enolpyruvylshikimate phosphate synthase, and
- 3-phosphoshikimate 1-carboxyvinyl transferase.
Function
The enzyme participates in biosynthesis of the aromatic amino acids phenylalanine, tyrosine and tryptophan. The enzyme is a target for herbicides as these amino acids are only synthesized in plants and microorganisms. Glyphosate acts as a competitive inhibitor for phosphoenolpyruvate and is used as a broad-spectrum systemic herbicide.[2][3]
Shikimate pathway
The shikimate pathway is a seven step metabolic route used by bacteria, fungi, and plants for the biosythesis of aromatic amino acids (phenylalanine, tyrosine, and tryptophan). This pathway is not found in animals, hence the products of this pathway represent essential amino acids that must be obtained from the animal's diet, with the exception of tyrosine, which mammals are capable of synthesizing from phenylalanine.
Structure
EPSP synthase is a monomeric enzyme. It is composed of two domains, which are joined by protein strands. This strand acts as a hinge, and can bring the two protein domains closer together. When a substrate binds to the enzyme, ligand bonding causes the two parts of the enzyme to clamp down around the substrate in the active site.
Reaction
EPSP synthase catalyzes the reaction which converts shikimate-3-phosphate plus phosphoenolpyruvate to 5-enolpyruvylshikimate-3-phosphate (EPSP).

Applications
Herbicides
Glyphosate is a chemical herbicide which kills plants by inhbiting the shikimate pathway. It targets EPSP synthase, the enzyme that catalyzes the conversion of shikimate-3-phosphate and phosphoenolpyruvate into EPSP. Glyphosate is a competitive inhibitor of the enzyme. Glyphosate resembles the transition state that transforms the reactants into products in the reaction that is catalyzed by EPSP synthase. Hence glyphosate (as a transition state analog) binds more tightly to EPSP synthase than its natural substrate and thereby prevents binding of substrate to the enzyme.[4]
This binding leads to inhibition of the enzyme and shuts down the entire pathway. Eventually this results in plant death from lack of aromatic amino acids used to make pigments and flavonoids the plant requires to survive. Glyphosate does not inhibit aromatic acid synthesis in animals and humans as they lack the shikimate pathway and aromatic amino acids are obtained from their diet.[4]
A version of the enzyme that both was resistant to glyphosate and that was still efficient enough to drive adequate plant growth was identified by Monsanto scientists after much trial and error in an Agrobacterium strain called CP4, which was found surviving in a waste-fed column at a glyphosate production facility; this version of enzyme, CP4 EPSPS, is the one that has been engineered into several genetically modified crops.[4][5]
References
- ↑ Priestman MA, Healy ML, Funke T, Becker A, Schönbrunn E (October 2005). "Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate". FEBS Lett. 579 (25): 5773–80. doi:10.1016/j.febslet.2005.09.066. PMID 16225867.
- ↑ Schönbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JN, Kabsch W (February 2001). "Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail". Proc. Natl. Acad. Sci. U.S.A. 98 (4): 1376–80. doi:10.1073/pnas.98.4.1376. PMC 29264. PMID 11171958.
- ↑ Pollegioni L, Schonbrunn E, Siehl D (August 2011). "Molecular basis of glyphosate resistance-different approaches through protein engineering". FEBS J. 278 (16): 2753–66. doi:10.1111/j.1742-4658.2011.08214.x. PMC 3145815. PMID 21668647.
- ↑ 4.0 4.1 4.2 Pollegioni L et al. Molecular basis of glyphosate resistance-different approaches through protein engineering. FEBS J. 2011 Aug;278(16):2753-66. PMID 21668647 PMC 3145815
- ↑ Green JM et al. Herbicide-resistant crops: utilities and limitations for herbicide-resistant weed management. J Agric Food Chem. 2011 Jun 8;59(11):5819-29. PMID 20586458 PMC 3105486
Further reading
- Morell H, Clark MJ, Knowles PF, Sprinson DB (1967). "The enzymic synthesis of chorismic and prephenic acids from 3-enolpyruvylshikimic acid 5-phosphate". J. Biol. Chem. 242 (1): 82–90. PMID 4289188.