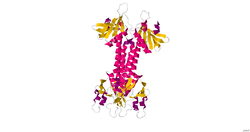ELMO1
Engulfment and cell motility protein 1 is a protein that in humans is encoded by the ELMO1 gene.[2][3] ELMO1 is located on chromosome number seven in humans and is located on chromosome number thirteen in mice.
Structure
The human engulfment and cell motility protein 1, ELMO1, is 720 residues in length. The protein contains the following three domains:
- N-terminal Armadillo domain (residues 82-262)
- central ELMO (Engulfment and Cell Motility) domain (301-492)
- C-terminal pleckstrin homology domain (residues 527-674)
ELMO1 also has a pro-rich motif at the extreme C terminus. Secondary structure analysis has predicted that there are alpha-helical regions at both the N and C-terminus.[1]
The structure of the pleckstrin homology domain of ELMO1 has been determine by X-ray crystallography.[1]
Function
The protein encoded by this gene interacts with the dedicator of cyto-kinesis 1 protein to promote phagocytosis and effect cell shape changes. Similarity to a C. elegans protein suggests that this protein may function in apoptosis and in cell migration. Alternative splicing of this gene results in multiple transcript variants encoding different isoforms.[3]
Interactions
ELMO1 has been shown to interact with Dock180[2][4] and HCK. ELMO1 directly interacts with the SH3 domain of HCK. The association between ELMO1 and HCK is dependent on polyproline interactions.[5]
When ELMO1 is complexed with DOCK180, Rac GTPase-dependent biological processes are activated. The pH domain of ELMO1 functions in trans to stabilize DOCK180 and make it resistant to degradation. When ELMO1 binds to DOCK180 it relieves the steric inhibition of DOCK180 which then activates the Rac GTPase. The pro-rich motif of the C terminus on ELMO1 is essential for the binding of ELMO1 to the SH3 domain at the N terminus of DOCK180.[1] The complex of ELMO1 and DOCK180 act as a regulator of Rac during development of a cell and cell migration. Mutation of both interaction sites for DOCK180 on ELMO1 will lead to the disruption of the ELMO1-DOCK180 complex. ELMO1 complexed with both DOCK180 and CrkII leads to maximal efficiency of phagocytosis in the cell. This complex of molecules happens upstream of Rac during phagocytosis. [2]
References
- ↑ 1.0 1.1 1.2 1.3 PDB 2VSZ; Komander D, Patel M, Laurin M, Fradet N, Pelletier A, Barford D, Côté JF (November 2008). "An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling". Mol. Biol. Cell 19 (11): 4837–51. doi:10.1091/mbc.E08-04-0345. PMC 2575150. PMID 18768751.
- ↑ 2.0 2.1 2.2 Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS (Oct 2001). "CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration". Cell 107 (1): 27–41. doi:10.1016/S0092-8674(01)00520-7. PMID 11595183.
- ↑ 3.0 3.1 "Entrez Gene: ELMO1 engulfment and cell motility 1".
- ↑ Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS (August 2002). "Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex". Nat. Cell Biol. 4 (8): 574–82. doi:10.1038/ncb824. PMID 12134158.
- ↑ Scott MP, Zappacosta F, Kim EY, Annan RS, Miller WT (August 2002). "Identification of novel SH3 domain ligands for the Src family kinase Hck. Wiskott-Aldrich syndrome protein (WASP), WASP-interacting protein (WIP), and ELMO1". J. Biol. Chem. 277 (31): 28238–46. doi:10.1074/jbc.M202783200. PMID 12029088.
Further reading
- Ohara O, Nagase T, Ishikawa K et al. (1997). "Construction and characterization of human brain cDNA libraries suitable for analysis of cDNA clones encoding relatively large proteins.". DNA Res. 4 (1): 53–9. doi:10.1093/dnares/4.1.53. PMID 9179496.
- Wiemann S, Weil B, Wellenreuther R et al. (2001). "Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs.". Genome Res. 11 (3): 422–35. doi:10.1101/gr.GR1547R. PMC 311072. PMID 11230166.
- Scott MP, Zappacosta F, Kim EY et al. (2002). "Identification of novel SH3 domain ligands for the Src family kinase Hck. Wiskott-Aldrich syndrome protein (WASP), WASP-interacting protein (WIP), and ELMO1.". J. Biol. Chem. 277 (31): 28238–46. doi:10.1074/jbc.M202783200. PMID 12029088.
- Brugnera E, Haney L, Grimsley C et al. (2002). "Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex.". Nat. Cell Biol. 4 (8): 574–82. doi:10.1038/ncb824. PMID 12134158.
- Strausberg RL, Feingold EA, Grouse LH et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Scherer SW, Cheung J, MacDonald JR et al. (2003). "Human chromosome 7: DNA sequence and biology.". Science 300 (5620): 767–72. doi:10.1126/science.1083423. PMC 2882961. PMID 12690205.
- Sanui T, Inayoshi A, Noda M et al. (2003). "DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1.". Blood 102 (8): 2948–50. doi:10.1182/blood-2003-01-0173. PMID 12829596.
- Katoh H, Negishi M (2003). "RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo.". Nature 424 (6947): 461–4. doi:10.1038/nature01817. PMID 12879077.
- Grimsley CM, Kinchen JM, Tosello-Trampont AC et al. (2004). "Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration.". J. Biol. Chem. 279 (7): 6087–97. doi:10.1074/jbc.M307087200. PMID 14638695.
- Wang X, Wu YC, Fadok VA et al. (2003). "Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12.". Science 302 (5650): 1563–6. doi:10.1126/science.1087641. PMID 14645848.
- Ota T, Suzuki Y, Nishikawa T et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs.". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Janardhan A, Swigut T, Hill B et al. (2006). "HIV-1 Nef binds the DOCK2-ELMO1 complex to activate rac and inhibit lymphocyte chemotaxis.". PLoS Biol. 2 (1): E6. doi:10.1371/journal.pbio.0020006. PMC 314466. PMID 14737186.
- Lu M, Kinchen JM, Rossman KL et al. (2004). "PH domain of ELMO functions in trans to regulate Rac activation via Dock180.". Nat. Struct. Mol. Biol. 11 (8): 756–62. doi:10.1038/nsmb800. PMID 15247908.
- Gerhard DS, Wagner L, Feingold EA et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC).". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- deBakker CD, Haney LB, Kinchen JM et al. (2005). "Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO.". Curr. Biol. 14 (24): 2208–16. doi:10.1016/j.cub.2004.12.029. PMID 15620647.
- Akakura S, Kar B, Singh S et al. (2005). "C-terminal SH3 domain of CrkII regulates the assembly and function of the DOCK180/ELMO Rac-GEF.". J. Cell. Physiol. 204 (1): 344–51. doi:10.1002/jcp.20288. PMID 15700267.
- Lu M, Kinchen JM, Rossman KL et al. (2005). "A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs.". Curr. Biol. 15 (4): 371–7. doi:10.1016/j.cub.2005.01.050. PMID 15723800.
- Shimazaki A, Kawamura Y, Kanazawa A et al. (2005). "Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy.". Diabetes 54 (4): 1171–8. doi:10.2337/diabetes.54.4.1171. PMID 15793258.
- Yokoyama N, deBakker CD, Zappacosta F et al. (2005). "Identification of tyrosine residues on ELMO1 that are phosphorylated by the Src-family kinase Hck.". Biochemistry 44 (24): 8841–9. doi:10.1021/bi0500832. PMC 2441568. PMID 15952790.