Demethylase
Demethylases are enzymes that remove methyl (CH3-) groups from nucleic acids, proteins (in particular histones), and other molecules. Demethylase enzymes are important in epigenetic modification mechanisms. The demethylase proteins alter transcriptional regulation of the genome by controlling the methylation levels that occur on DNA and histones and, in turn, regulate the chromatin state at specific gene loci within organisms.
Oxidative demethylation
Histone demethylation
For many years histone methylation was thought to be irreversible, due to the fact that the half-life of the histone methylation was approximately equal to the half-life of histones themselves.[1] In 2004, Shi et al. published their discovery of the histone demethylase LSD1 (later classified as KDM1A), a nuclear amine oxidase homolog.[2] With the new found interest in epigenetics, many more families of histone demethylases have been found. Defined by their mechanisms, two main classes of histone demethylases exist: a flavin adenine dinucleotide (FAD)-dependent amine oxidase, and an Fe(II) and α-ketoglutarate-dependent dioxygenase, with the general proposed mechanisms shown in the figure below.[3]

Histone demethylase proteins have a variety of domains that serve different functions. These functions include binding to the histone (or sometimes the DNA on the nucleosome), recognizing the correct methylated amino acid substrate and catalyzing the reaction, and binding cofactors. Cofactors include: alpha-keto glutarate (JmjC-domain containing demethylases), CoREST (LSD), FAD, Fe (II) or NOG (N-oxalylglycine).[4] Domains include:
- SWIRM1 (Swi3, Rsc, and Moira domain): Proposed anchor site for histone molecules; found in several chromatin modifying complexes; facilitates demethylase protein and co-factor CoREST (nucleosomal substrate binding)[5]
- Jumonji (N/C terminal domains): Binding domain of key cofactors such as alpha-keto glutarate; connected by a beta-hairpin/mixed domain[4]
- PHD-finger: hydrophobic cage of residues that acts to bind methylated peptides; plays key role in recognition and selectivity for methylated histone residues[4]
- Zinc-finger: DNA binding domain[4]
- Amine oxidase domain: catalytic active site of LSD proteins; larger than related proteins to help fit size of the histone tail [4]
_with_domains_highlighted.png)

There are several families of histone demethylases, which act on different substrates and play different roles in cellular function. A code has been developed to indicate the substrate for a histone demethylase. The substrate is first specified by the histone subunit (H1, H2A, H2B, H3, H4) and then the one letter designation and number of the amino acid that is methylated. Lastly, the level of methylation is sometimes noted by the addition of "me#", with the numbers being 1, 2, and 3 for monomethylated, dimethylated, and trimethylated substrates, respectively. For example, H3K9me2 is histone H3 with a dimethylated lysine in the ninth position.
- KDM1
- The KDM1 family includes KDM1A and KDM1B. KDM1A (also referred to as LSD1/AOF2/BHC110) can act on mono- and dimethylated H3K4 and H3K9, and KDM1B (also referred to as LSD2/AOF1) acts only on mono- and dimethylated H3K4. These enzymes can have roles critical in embryogenesis and tissue-specific differentiation, as well as oocyte growth.[1] KDM1A was the first demethylase to be discovered and thus it has been studied most extensively.[2]
- Deletion of the gene for KDM1A can have effects on the growth and differentiation of embryonic stem cells and can lead to embryonic lethality in knockout mice, who do not produce the KDM1A gene product.[6][7] KDM1A is also thought to play a role in cancer, as poorer outcomes can be correlated with higher expression of this gene.[8][9] Therefore, the inhibition of KDM1A may be a possible treatment for cancer.[10][11] KDM1A has many different binding partners, which may be necessary for its demethylation activity.[12]
- KDM1B, however, is mostly involved in oocyte development. Deletion of this gene leads to maternal effect lethality in mice.[13] Orthologs of KDM1 in D. melanogaster and C. elegans appear to function similarly to KDM1B rather than KDM1A.[14][15]
- KDM2
- The KDM2 family includes KDM2A and KDM2B. KDM2A (also referred to as JHDM1A/FBXL11) can act on mono- and dimethylated H3K36 and trimethylated H3K4. KDM2B (also referred to as JHDM1B/FBXL10) acts only on mono- and dimethylated H3K36. KDM2A has roles in either promoting or inhibiting tumor function, and KDM2B has roles in oncogenesis.[1]
- In many eukaryotes, the KDM2A protein contains a CXXC zinc finger domain capable of binding unmethylated CpG islands. It is currently thought that KDM2A proteins may bind to many gene regulatory elements without the aid of sequence specific transcription factors.[16] Although the role of KDM2 in eukaryotic developmental differentiation is still largely a mystery, both KDM2A and KDM2B have been shown to play roles in tumor growth and suppression. KDM2B has been shown to be over-expressed in human lymphomas and adenocarcinomas; prostate cancers and glioblastomas, however, show reduced expression of both KDM2A and KDM2B. Additionally, KDM2B has been shown to prevent senescence in some cells through ectopic expression further indicating its potential as an oncogene.[17]
- KDM3
- The KDM3 family includes KDM3A, KDM3B and JMJD1C. KDM3A (also referred to as JHDM2A/JMJD1A/TSGA) can act on mono- and dimethylated H3K9. The substrates for KDM3B (also referred to as JHDM2B/JMJD1B) and JMJD1C (also referred to as JHDM2C/TRIP8) are not known.[18] The KDM3A has roles in spermatogenesis and metabolic functions; the roles are of KDM3B and JMJD1C are unknown.[1]
- Knockdown studies of KDM3A in mice, where the mouse produces reduced levels of KDM3A, resulted in male infertility and adult onset-obesity. Additional studies have indicated that KDM3A may play a role in regulation of androgen receptor-dependent genes as well as genes involved in pluripotency, indicating a potential role for KDM3A in tumorigenesis.[19]
- KDM4
- The KDM4 family includes KDM4A, KDM4B, KDM4C, and KDM4D. These are also referred to as JMDM3A/JMJD2A, JMDM3B/JMJD2B, JMDM3C/JMJD2C, and JMDM3D/JMJD2D, respectively. These enzymes can act on di- and trimethylated H3K9, H3K36, H1K26. KDM4B and KDM4C have roles in tumorigenesis, and the role of KDM4D is unknown.[1]
- The KDM4 family of proteins have been linked to malignant transformation. Specifically, KDM4C amplification has been documented in oesophageal squamous carcinomas, medulloblastomas and breast cancers; amplification of KDM4B has also been found in medulloblastomas.[20][21][22][23] Other gene expression data has also suggested KDM4A, KDM4B, and KDM4C are over-expressed in prostate cancer.[24]
- KDM5
- The KDM5 family includes KDM5A, KDM5B, KDM5C, and KDM5D. These are also referred to as JARID1A/RBP2, JARID1B/PLU-1, JARID1C/SMCX, and JARID1D/SMCY, respectively. These enzymes can act on di- and trimethylated H3K4.[1]
- KDM5 protein family appear to play key developmental functions. The deletion of the JmjC domain of retinoblastoma binding protein related 2 (RBR-2) in C. elegans express defects in vulva formation.[25] Mutations to the JmjC domain in Drosophila causes either lethal effects on larval or many developmental defects in those that survive.[26]
- KDM5A in cell culture systems have also shown links to regulation of differentiation, mitochondrial function, cell cycle progression.[27][28][29][30][31][32] KDM5B and KDM5C have also shown to interaction with PcG proteins, which are involved in transcriptional repression. KDM5C mutations (found on the X-chromosome) have also been found in patients with X-linked mental retardation.[33] Depletion of KDM5C homologs in D. rerio have shown brain-patterning defects and neuronal cell death.[34]
- KDM6
- The KDM6 family includes KDM6A, KDM6B, and UTY. KDM6A (also referred to as UTX) and KDM6B (also referred to as JMJD3) act on di- and trimethylated H3K27 and have roles in development; the substrate and role of UTY is unknown.[1] On the whole, both KDM6A and KDM6B possess tumor-suppressive characteristics. KDM6A knockdowns in fibroblasts lead to an immediate increase in fibroblast population. KDM6B expressed in fibroblasts induces oncogenes of the RAS_RAF pathway.[35] Deletions and point mutations of KDM6A have been identified as one cause of Kabuki Syndrome, a congenital disorder resulting in intellectual disability.[36][37]
- Other possible roles have been suggested for KDM6B. Specifically in one study, mutating homologs of KDM6B disrupted gonadal development in C.elegans.[38] Other studies have shown that KDM6B expression is up-regulated in activated macrophages and dynamically expressed during differentiation of stem cells.[39][40]
- On the other hand, depletion of homologs of KDM6A in D. rerio have shown decreased expression of HOX genes, which play a role in regulating body patterning during development.[41] In mammalian studies, KDM6A has been shown to regulate HOX genes as well.[38][42]
Ester Demethylation

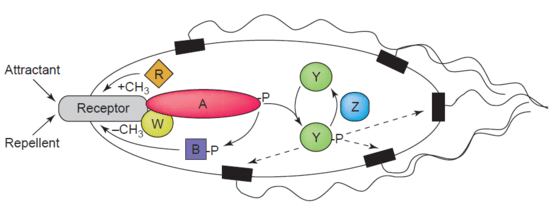
Another example of a demethylase is protein-glutamate methylesterase, also known as CheB protein (EC 3.1.1.61), which demethylates MCPs (methyl-accepting chemotaxis proteins) through hydrolysis of carboxylic ester bonds. The association of a chemotaxis receptor with an agonist leads to the phosphorylation of CheB. Phosphorylation of CheB protein enhances its catalytic MCP demethylating activity resulting in adaption of the cell to environmental stimuli.[43] MCPs respond to extracellular attractants and repellents in bacteria like E. coli in chemotaxis regulation. CheB is more specifically termed a methylesterase, as it removes methyl groups from methylglutamate residues located on the MCPs through hydrolysis, producing glutamate accompanied by the release of methanol.[44]
CheB is of particular interest to researchers as it may be a therapeutic target for mitigating the spread of bacterial infections.[45]
See also
- Esterase
- Transferase
- Methylase
- Chemotaxis
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Pedersen, M.T. et al. "Histone Demethylases in Development and Disease" Cell Trends in Cell Biology. Nov, 2010;20,11:662-71. {{doi:http://dx.doi.org/10.1016/j.tcb.2010.08.011}}
- ↑ 2.0 2.1 Shi, Y., et al. "Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1" Cell. Dec 29, 2004;119:941-53. {{doi:10.1016/j.cell.2004.12.012}}
- ↑ Klose RJ & Zhang Y. 2007. "Regulation of histone methylation by demethylimination and demethylation" Nature Reviews Mol. Cell Biology. April 2007;8:307-18. {{doi:http://dx.doi.org/10.1038/nrm2143}}
- ↑ 4.0 4.1 4.2 4.3 4.4 Mosamaparast, Nima, et al. "Reversal of Histone Methylation: Biochemical and Molecular Mechanisms of Histone Demethylases." Annu. Rev. Biochem. 79.(2010): 155.
- ↑ Tochio, Naoya, et al. "Solution Structure of the SWIRM Domain of Human Histone Demethylase LSD1." Structure 14.3 (2006):457.
- ↑ Wang, J. et al. (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129
- ↑ Wang, J. et al. (2007) Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446, 882–887
- ↑ Kahl, P. et al. (2006) Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 66, 11341– 11347
- ↑ Lim, S. et al. (2010) Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31, 512– 20
- ↑ Metzger, E. et al. (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439
- ↑ Schulte, J.H. et al. (2009) Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 69, 2065–2071
- ↑ Wang, Y. et al. (2009) LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660–672
- ↑ Ciccone, D.N. et al. (2009) KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418
- ↑ Rudolph, T. et al. (2007) Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol. Cell 26, 103–115
- ↑ Di Stefano, L., Ji, J.Y., Moon, N.S., Herr, A. and Dyson, N. (2007) Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr. Biol. 17, 808–812
- ↑ Blackledge, N.P. et al. (2010) CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell 38, 179–190
- ↑ He, J. et al.(2008) The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat. Struct. Mol. Biol. 15, 1169–1175
- ↑ Yamane, K. et al. (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495
- ↑ Loh, Y.H. et al. (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557
- ↑ Ehrbrecht, A. et al. (2006) Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J. Pathol. 208, 554–563
- ↑ Liu, G. et al. (2009) Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 28, 4491–4500
- ↑ Northcott, P.A. et al. (2009) Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat. Genet. 41, 465–472
- ↑ Yang, Z.Q. et al. (2000) Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 60, 4735–4739
- ↑ Cloos, P.A. et al. (2006) The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442, 307–311
- ↑ Christensen, J. et al. (2007) RBP2 belongs to a family of demethylases, specific for tri- and dimethylated lysine 4 on histone 3. Cell 128, 1063–1076
- ↑ Gildea, J.J. et al. (2000) A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156, 645–663
- ↑ Lee, N. et al. (2009) The H3K4 demethylase lid associates with and inhibits histone deacetylase Rpd3. Mol. Cell Biol. 29, 1401–1410
- ↑ Benevolenskaya, E.V. et al. (2005) Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18, 623–635
- ↑ Lopez-Bigas, N. et al. (2008) Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol. Cell 31, 520–530
- ↑ Pasini, D. et al. (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 22, 1345–1355
- ↑ van Oevelen, C. et al. (2008) A role for mammalian Sin3 in permanentgene silencing. Mol. Cell 32, 359–370
- ↑ Zeng, J. et al. (2010) The histone demethylase RBP2 Is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology 138, 981–992
- ↑ Jensen, L.R. et al. (2005) Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236
- ↑ Iwase, S. et al. (2007) The X-linked mental retardation gene SMCX/ JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088
- ↑ Agger, K. et al. "The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence" Genes Dev. 2009;23:1171–1176
- ↑ Lederer, D., Grisart, B., Digilio, M. C., Benoit, V., Crespin, M., Ghariani, S. C., ... & Verellen-Dumoulin, C. (2012). Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. The American Journal of Human Genetics, 90(1), 119-124.
- ↑ Miyake, Noriko, et al. "KDM6A point mutations cause Kabuki syndrome." Human mutation 34.1 (2013): 108-110.
- ↑ 38.0 38.1 Agger, K. et al. "UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development" Nature. 2007;449:731–734
- ↑ De Santa, F. et al. "The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing" Cell. 2007;130:1083–1094
- ↑ Burgold, T. et al. (2008) The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 3, e3034
- ↑ Lan, F. et al. "A histone H3 lysine 27 demethylase regulates animal posterior development" Nature. 2007;449:689–694
- ↑ Wang, J.K. et al. (2010) The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 24, 327–332
- ↑ 43.0 43.1 Vladimirov, N., et al. "Dependence of bacterial chemotaxis on gradient shape and adaptation rate." PLoS computational biology 4.12 (2008): e1000242.
- ↑ Park, B., et al. "Reconstruction of the chemotaxis receptor-kinase assembly" Nature Structural & Molecular Biology 13, 400-407 (2006). 23 April 2006
- ↑ West, Ann H., Erik Martinez-Hackert, and Ann M. Stock. "Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB." Journal of molecular biology 250.2 (1995): 276.