Defective Interfering RNA
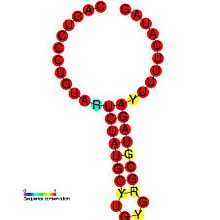
In virology, defective interfering RNAs (DI-RNA) are non-infectious, mutated subviral RNAs that play a role in the evolution of viral diseases. DI-RNAs contain viral structural proteins but are missing a part of the virus's genome. They are naturally occurring and can be synthesized for experimental use. They are spontaneously produced by error-prone viral replicase, or RNA-dependent RNA polymerase during viral replication.[1][2] They have defective interfering particles that are replication defective but can infect host cells along with a normal virus's help. DI-RNA replication is regulated by the Coronavirus SL-III cis-acting replication element (shown in the image) and coronaviruses have been shown to be important in persistent infections.[3] A more in-depth introduction can be found in Alice Huang and David Baltimore's work from 1970.[4]
Defection
The RNAs are considered defective when they lack the genes for independent replication and may also lack movement/encapsulation leaving them dependent on the parent RNA and its byproducts to function.[1] They usually retain the ends of non-defective RNA[5] but never retain their own replicase. Even though they are missing parts of the genome, DI-RNAs can still replicate with aid from a helper virus and replication signals.[6] [7]
Types of defective genomes
- Deletions defections are when a fragment of the template is skipped. Examples of this type of defection can be found in the Resende, et al. 1992 article.[5]
- Snapbacks defections are when replicase transcribes part of one strand then uses this new strand as a template. The result of this can produce a hairpin. Snapback defections are explored in the Schubert & Lazzarini 1981 article.[8]
- Panhandle defections are when the polymerase carries a partially made strand and then switches back to transcribe the 5' end, forming the panhandle shape. Panhandle defections in the influenza virus can be found in the Fodor, et al. 1994 article.[9]
- Compound defections are when both a deletion and snapback defection happens together.
Interference
The RNAs are considered interfering when they effect the function of the parent RNA through competitive inhibition.[1] In other words, defective and non-defective viruses replicate simultaneously, but when defective particles increase, the amount of replicated non-defective virus is decreased. The extent of interference depends on the type of defection in the genome. This interfering nature is becoming more and more important for future research on virus therapies.[10] It is thought that because of their specificity, DI-RNA will be targeted to sites of infection. There, scientists have created "protecting viruses" made up of RNA containing DI-RNAs and are working to implement them in hopes to reduce the amount of infection.[11]
Recent published work
Recent work has been done by virologists to learn more about the interference in infection of host cells and how DI-RNAs could potentially work as antiviral agents.[12] The Dimmock & Easton, 2014 article explains that pre-clinical work is being done to test their effectiveness against influenza viruses.[13] DI-RNAs have also been found to aid in the infection of fungi via viruses of the family Partitiviridae for the first time, which makes room for more interdisciplinary work.[2]
References
- ↑ 1.0 1.1 1.2 Pathak, K. B.; Nagy, P. D. (2009). "Defective Interfering RNAs: Foes of Viruses and Friends of Virologists". Viruses 1 (3): 895. doi:10.3390/v1030895.
- ↑ 2.0 2.1 Chiba, S; Lin, Y. H.; Kondo, H; Kanematsu, S; Suzuki, N (2013). "Effects of defective interfering RNA on symptom induction by, and replication of, a novel partitivirus from a phytopathogenic fungus, Rosellinia necatrix". Journal of Virology 87 (4): 2330–41. doi:10.1128/JVI.02835-12. PMC 3571465. PMID 23236074.
- ↑ Siddell, S; Wege, H; Ter Meulen, V (1982). "The structure and replication of coronaviruses". Current topics in microbiology and immunology 99: 131–63. PMID 7047085.
- ↑ Huang, A. S.; Baltimore, D (1970). "Defective viral particles and viral disease processes". Nature 226 (5243): 325–7. PMID 5439728.
- ↑ 5.0 5.1 Resende Rde, O; De Haan, P; Van De Vossen, E; De Avila, A. C.; Goldbach, R; Peters, D (1992). "Defective interfering L RNA segments of tomato spotted wilt virus retain both virus genome termini and have extensive internal deletions". The Journal of general virology. 73 ( Pt 10): 2509–16. PMID 1402797.
- ↑ Makino, S; Shieh, C. K.; Soe, L. H.; Baker, S. C.; Lai, M. M. (1988). "Primary structure and translation of a defective interfering RNA of murine coronavirus". Virology 166 (2): 550–60. PMID 2845661.
- ↑ Palmer, S.R. (15 September 2011). Oxford Textbook of Zoonoses: Biology, Clinical Practice, and Public Health Control (2nd Edition ed.). Oxford University Press. pp. 399–400.
- ↑ Schubert, M; Lazzarini, R. A. (1981). "Structure and origin of a snapback defective interfering particle RNA of vesicular stomatitis virus". Journal of virology 37 (2): 661–72. PMC 171054. PMID 6261012.
- ↑ Fodor, E; Pritlove, D. C.; Brownlee, G. G. (1994). "The influenza virus panhandle is involved in the initiation of transcription". Journal of virology 68 (6): 4092–6. PMC 236924. PMID 8189550.
- ↑ Thompson, K. A.; Yin, J (2010). "Population dynamics of an RNA virus and its defective interfering particles in passage cultures". Virology Journal 7: 257. doi:10.1186/1743-422X-7-257. PMC 2955718. PMID 20920247.
- ↑ Easton, A. J.; Scott, P. D.; Edworthy, N. L.; Meng, B; Marriott, A. C.; Dimmock, N. J. (2011). "A novel broad-spectrum treatment for respiratory virus infections: Influenza-based defective interfering virus provides protection against pneumovirus infection in vivo". Vaccine 29 (15): 2777–84. doi:10.1016/j.vaccine.2011.01.102. PMID 21320545.
- ↑ Marriott, A. C.; Dimmock, N. J. (2010). "Defective interfering viruses and their potential as antiviral agents". Reviews in Medical Virology 20 (1): 51–62. doi:10.1002/rmv.641. PMID 20041441.
- ↑ Dimmock, N. J.; Easton, A. J. (2014). "Defective interfering influenza virus RNAs: Time to reevaluate their clinical potential as broad-spectrum antivirals?". Journal of Virology 88 (10): 5217–27. doi:10.1128/JVI.03193-13. PMC 4019098. PMID 24574404.
| ||||||||||||||||||||||||||||||