D-xylulose reductase
| D-xylulose reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.1.1.9 | ||||||||
| CAS number | 9028-16-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
In enzymology, a D-xylulose reductase (EC 1.1.1.9) is an enzyme that catalyzes the chemical reaction
- xylitol + NAD+
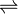 D-xylulose + NADH + H+
D-xylulose + NADH + H+
Thus, the two substrates of this enzyme are xylitol and NAD+, whereas its 3 products are D-xylulose, NADH, and H+.
This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is xylitol:NAD+ 2-oxidoreductase (D-xylulose-forming). Other names in common use include NAD+-dependent xylitol dehydrogenase, xylitol dehydrogenase, erythritol dehydrogenase, 2,3-cis-polyol(DPN) dehydrogenase (C3-5), pentitol-DPN dehydrogenase, and xylitol-2-dehydrogenase. This enzyme participates in pentose and glucuronate interconversions.
Structural studies
As of late 2007, only one structure has been solved for this class of enzymes, with the PDB accession code 1ZEM.
References
- Chiang C, Knight SG (1960). "A new pathway of pentose metabolism". Biochem. Biophys. Res. Commun. 3 (5): 554–9. doi:10.1016/0006-291X(60)90174-1. PMID 13692998.
- Hickman J and Ashwell G (1959). "A sensitive and stereospecific enzymatic assay for xylulose". J. Biol. Chem. 234: 758–761.
- Jakoby WB, Fredericks J (1961). "Erythritol dehydrogenase from Aerobacter aerogenes". Biochim. Biophys. Acta 48: 26–32. doi:10.1016/0006-3002(61)90511-X. PMID 13789254.