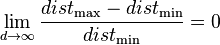Clustering high-dimensional data
Clustering high-dimensional data is the cluster analysis of data with anywhere from a few dozen to many thousands of dimensions. Such high-dimensional data spaces are often encountered in areas such as medicine, where DNA microarray technology can produce a large number of measurements at once, and the clustering of text documents, where, if a word-frequency vector is used, the number of dimensions equals the size of the vocabulary.
Problems
Four problems need to be overcome for clustering in high-dimensional data:[1]
- Multiple dimensions are hard to think in, impossible to visualize, and, due to the exponential growth of the number of possible values with each dimension, complete enumeration of all subspaces becomes intractable with increasing dimensionality. This problem is known as the curse of dimensionality.
- The concept of distance becomes less precise as the number of dimensions grows, since the distance between any two points in a given dataset converges. The discrimination of the nearest and farthest point in particular becomes meaningless:
- A cluster is intended to group objects that are related, based on observations of their attribute's values. However, given a large number of attributes some of the attributes will usually not be meaningful for a given cluster. For example, in newborn screening a cluster of samples might identify newborns that share similar blood values, which might lead to insights about the relevance of certain blood values for a disease. But for different diseases, different blood values might form a cluster, and other values might be uncorrelated. This is known as the local feature relevance problem: different clusters might be found in different subspaces, so a global filtering of attributes is not sufficient.
- Given a large number of attributes, it is likely that some attributes are correlated. Hence, clusters might exist in arbitrarily oriented affine subspaces.
Recent research indicates that the discrimination problems only occur when there is a high number of irrelevant dimensions, and that shared-nearest-neighbor approaches can improve results.[2]
Approaches
Approaches towards clustering in axis-parallel or arbitrarily oriented affine subspaces differ in how they interpret the overall goal, which is finding clusters in data with high dimensionality.[1] An overall different approach is to find clusters based on pattern in the data matrix, often referred to as biclustering, which is a technique frequently utilized in bioinformatics.
Subspace clustering

Subspace clustering is the task of detecting all clusters in all subspaces. This means that a point might be a member of multiple clusters, each existing in a different subspace. Subspaces can either be axis-parallel or affine. The term is often used synonymously with general clustering in high-dimensional data.
The image on the right shows a mere two-dimensional space where a number of clusters can be identified. In the one-dimensional subspaces, the clusters 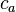 (in subspace
(in subspace 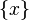 ) and
) and 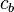 ,
,  ,
, 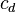 (in subspace
(in subspace  ) can be found.
) can be found.  cannot be considered a cluster in a two-dimensional (sub-)space, since it is too sparsely distributed in the
cannot be considered a cluster in a two-dimensional (sub-)space, since it is too sparsely distributed in the 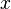 axis. In two dimensions, the two clusters
axis. In two dimensions, the two clusters 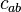 and
and 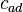 can be identified.
can be identified.
The problem of subspace clustering is given by the fact that there are 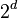 different subspaces of a space with
different subspaces of a space with 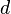 dimensions. If the subspaces are not axis-parallel, an infinite number of subspaces is possible. Hence, subspace clustering algorithm utilize some kind of heuristic to remain computationally feasible, at the risk of producing inferior results. For example, the downward-closure property (cf. association rules) can be used to build higher-dimensional subspaces only by combining lower-dimensional ones, as any subspace T containing a cluster, will result in a full space S also to contain that cluster (i.e. S ⊆ T), an approach taken by most of the traditional algorithms such as CLIQUE,[3] SUBCLU.[4] It is also possible to define a subspace using different degrees of relevance for each dimension, an approach taken by iMWK-Means.[5]
dimensions. If the subspaces are not axis-parallel, an infinite number of subspaces is possible. Hence, subspace clustering algorithm utilize some kind of heuristic to remain computationally feasible, at the risk of producing inferior results. For example, the downward-closure property (cf. association rules) can be used to build higher-dimensional subspaces only by combining lower-dimensional ones, as any subspace T containing a cluster, will result in a full space S also to contain that cluster (i.e. S ⊆ T), an approach taken by most of the traditional algorithms such as CLIQUE,[3] SUBCLU.[4] It is also possible to define a subspace using different degrees of relevance for each dimension, an approach taken by iMWK-Means.[5]
Projected clustering
Projected clustering seeks to assign each point to a unique cluster, but clusters may exist in different subspaces. The general approach is to use a special distance function together with a regular clustering algorithm.
For example, the PreDeCon algorithm checks which attributes seem to support a clustering for each point, and adjusts the distance function such that dimensions with low variance are amplified in the distance function.[6] In the figure above, the cluster  might be found using DBSCAN with a distance function that places less emphasis on the
might be found using DBSCAN with a distance function that places less emphasis on the 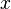 -axis and thus exaggerates the low difference in the
-axis and thus exaggerates the low difference in the 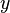 -axis sufficiently enough to group the points into a cluster.
-axis sufficiently enough to group the points into a cluster.
PROCLUS uses a similar approach with a k-medoid clustering.[7] Initial medoids are guessed, and for each medoid the subspace spanned by attributes with low variance is determined. Points are assigned to the medoid closest, considering only the subspace of that medoid in determining the distance. The algorithm then proceeds as the regular PAM algorithm.
If the distance function weights attributes differently, but never with 0 (and hence never drops irrelevant attributes), the algorithm is called a "soft"-projected clustering algorithm.
Hybrid approaches
Not all algorithms try to either find a unique cluster assignment for each point or all clusters in all subspaces; many settle for a result in between, where a number of possibly overlapping, but not necessarily exhaustive set of clusters are found. An example is FIRES, which is from its basic approach a subspace clustering algorithm, but uses a heuristic too aggressive to credibly produce all subspace clusters.[8]
Correlation clustering
Another type of subspaces is considered in Correlation clustering (Data Mining).
Software
- ELKI includes various subspace and correlation clustering algorithms
References
- ↑ 1.0 1.1 Kriegel, H. P.; Kröger, P.; Zimek, A. (2009). "Clustering high-dimensional data". ACM Transactions on Knowledge Discovery from Data 3: 1. doi:10.1145/1497577.1497578.
- ↑ Houle, M. E.; Kriegel, H. P.; Kröger, P.; Schubert, E.; Zimek, A. (2010). Can Shared-Neighbor Distances Defeat the Curse of Dimensionality? (PDF). Scientific and Statistical Database Management. Lecture Notes in Computer Science 6187. p. 482. doi:10.1007/978-3-642-13818-8_34. ISBN 978-3-642-13817-1.
- ↑ Agrawal, R.; Gehrke, J.; Gunopulos, D.; Raghavan, P. (2005). "Automatic Subspace Clustering of High Dimensional Data". Data Mining and Knowledge Discovery 11: 5. doi:10.1007/s10618-005-1396-1.
- ↑ Kailing, K.; Kriegel, H. P.; Kröger, P. (2004). Density-Connected Subspace Clustering for High-Dimensional Data. Proceedings of the 2004 SIAM International Conference on Data Mining. p. 246. doi:10.1137/1.9781611972740.23. ISBN 978-0-89871-568-2.
- ↑ Cordeiro De Amorim, R.; Mirkin, B. (2012). "Minkowski metric, feature weighting and anomalous cluster initializing in K-Means clustering". Pattern Recognition 45 (3): 1061. doi:10.1016/j.patcog.2011.08.012.
- ↑ Böhm, C.; Kailing, K.; Kriegel, H. -P.; Kröger, P. (2004). Density Connected Clustering with Local Subspace Preferences. Fourth IEEE International Conference on Data Mining (ICDM'04). p. 27. doi:10.1109/ICDM.2004.10087. ISBN 0-7695-2142-8.
- ↑ Aggarwal, C. C.; Wolf, J. L.; Yu, P. S.; Procopiuc, C.; Park, J. S. (1999). "Fast algorithms for projected clustering". ACM SIGMOD Record 28 (2): 61. doi:10.1145/304181.304188.
- ↑ Kriegel, H.; Kröger, P.; Renz, M.; Wurst, S. (2005). A Generic Framework for Efficient Subspace Clustering of High-Dimensional Data. Fifth IEEE International Conference on Data Mining (ICDM'05). p. 250. doi:10.1109/ICDM.2005.5. ISBN 0-7695-2278-5.