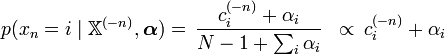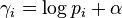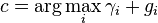Categorical distribution
| Parameters |
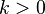 number of categories (integer) number of categories (integer)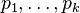 event probabilities ( event probabilities (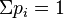 ) ) |
|---|---|
| Support |
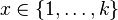 |
| pmf |
(1)
|
| CDF |
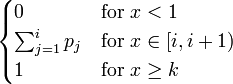 |
| Mean |
![\operatorname{E}([x=i]) = p_i](../I/m/070ddfd01477aa2c634850c84f6d87d9.png) , this is the mean of the Iverson bracket , this is the mean of the Iverson bracket ![[x=i]](../I/m/2bd4b55c9539155fb4b0da36be8424a1.png) and not the mean of and not the mean of 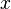 |
| Median |
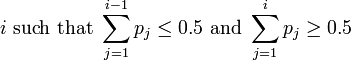 |
| Mode |
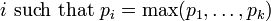 |
| Variance |
![\textstyle{\mathrm{Var}}([x=i]) = p_i (1-p_i)](../I/m/71274c135d41e4e7d6b25f66d284cbe1.png) ![\textstyle {\mathrm{Cov}}([x=i],[x=j]) = - p_i p_j~~(i\neq j)](../I/m/33afb8f81731ef5641596f019793907b.png) |
| MGF |
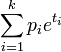 |
| CF |
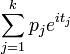 where where 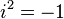 |
| PGF |
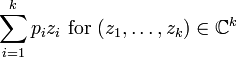 |
In probability theory and statistics, a categorical distribution (also called a "generalized Bernoulli distribution" or, less precisely, a "discrete distribution") is a probability distribution that describes the result of a random event that can take on one of K possible outcomes, with the probability of each outcome separately specified. There is not necessarily an underlying ordering of these outcomes, but numerical labels are attached for convenience in describing the distribution, often in the range 1 to K. Note that the K-dimensional categorical distribution is the most general distribution over a K-way event; any other discrete distribution over a size-K sample space is a special case. The parameters specifying the probabilities of each possible outcome are constrained only by the fact that each must be in the range 0 to 1, and all must sum to 1.
The categorical distribution is the generalization of the Bernoulli distribution for a categorical random variable, i.e. for a discrete variable with more than two possible outcomes.
Terminology
Occasionally, the categorical distribution is termed the "discrete distribution". However, this properly refers not to one particular family of distributions but to a general class of distributions.
Note that, in some fields, such as machine learning and natural language processing, the categorical and multinomial distributions are conflated, and it is common to speak of a "multinomial distribution" when a categorical distribution is actually meant.[1] This imprecise usage stems from the fact that it is sometimes convenient to express the outcome of a categorical distribution as a "1-of-K" vector (a vector with one element containing a 1 and all other elements containing a 0) rather than as an integer in the range 1 to K; in this form, a categorical distribution is equivalent to a multinomial distribution for a single observation (see below).
However, conflating the categorical and multinomial distributions can lead to problems. For example, in a Dirichlet-multinomial distribution, which arises commonly in natural language processing models (although not usually with this name) as a result of collapsed Gibbs sampling where Dirichlet distributions are collapsed out of a Hierarchical Bayesian model, it is very important to distinguish categorical from multinomial. The joint distribution of the same variables with the same Dirichlet-multinomial distribution has two different forms depending on whether it is characterized as a distribution whose domain is over individual categorical nodes or over multinomial-style counts of nodes in each particular category (similar to the distinction between a set of Bernoulli-distributed nodes and a single binomial-distributed node). Both forms have very similar-looking probability mass functions (PMF's), which both make reference to multinomial-style counts of nodes in a category. However, the multinomial-style PMF has an extra factor, a multinomial coefficient, that is a constant equal to 1 in the categorical-style PMF. Confusing the two can easily lead to incorrect results in settings where this extra factor is not constant with respect to the distributions of interest. The factor is frequently constant in the complete conditionals used in Gibbs sampling and the optimal distributions in variational methods.
Introduction
A categorical distribution is a discrete probability distribution whose sample space is the set of k individually identified items. It is the generalization of the Bernoulli distribution for a categorical random variable.
In one formulation of the distribution, the sample space is taken to be a finite sequence of integers. The exact integers used as labels are unimportant; they might be {0, 1, ..., k-1} or {1, 2, ..., k} or any other arbitrary set of values. In the following descriptions, we use {1, 2, ..., k} for convenience, although this disagrees with the convention for the Bernoulli distribution, which uses {0, 1}. In this case, the probability mass function f is:
where  ,
, 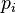 represents the probability of seeing element i and
represents the probability of seeing element i and 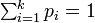 .
.
Another formulation that appears more complex but facilitates mathematical manipulations is as follows, using the Iverson bracket:[2]
where ![[x=i]](../I/m/2bd4b55c9539155fb4b0da36be8424a1.png) evaluates to 1 if
evaluates to 1 if 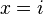 , 0 otherwise. There are various advantages of this formulation, e.g.:
, 0 otherwise. There are various advantages of this formulation, e.g.:
- It is easier to write out the likelihood function of a set of independent identically distributed categorical variables.
- It connects the categorical distribution with the related multinomial distribution.
- It shows why the Dirichlet distribution is the conjugate prior of the categorical distribution, and allows the posterior distribution of the parameters to be calculated.
Yet another formulation makes explicit the connection between the categorical and multinomial distributions by treating the categorical distribution as a special case of the multinomial distribution in which the parameter n of the multinomial distribution (the number of sampled items) is fixed at 1. In this formulation, the sample space can be considered to be the set of 1-of-K encoded[3] random vectors x of dimension k having the property that exactly one element has the value 1 and the others have the value 0. The particular element having the value 1 indicates which category has been chosen. The probability mass function f in this formulation is:
where 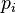 represents the probability of seeing element i and
represents the probability of seeing element i and  .
This is the formulation adopted by Bishop.[3][nb 1]
.
This is the formulation adopted by Bishop.[3][nb 1]
Properties
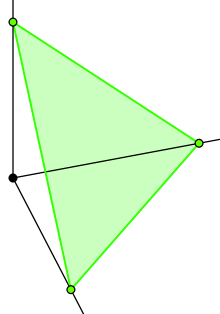
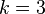 are the 2-simplex
are the 2-simplex  , embedded in 3-space.
, embedded in 3-space.- The distribution is completely given by the probabilities associated with each number i:
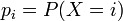 , i = 1,...,k, where
, i = 1,...,k, where  . The possible probabilities are exactly the
. The possible probabilities are exactly the 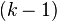 ; for k = 2 this reduces to the possible probabilities of the Bernoulli distribution being the 1-simplex,
; for k = 2 this reduces to the possible probabilities of the Bernoulli distribution being the 1-simplex, 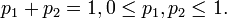
- The distribution is a special case of a "multivariate Bernoulli distribution"[4] in which exactly one of the k 0-1 variables takes the value one.
- Let
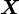 be the realisation from a categorical distribution. Define the random vector Y as composed of the elements:
be the realisation from a categorical distribution. Define the random vector Y as composed of the elements:
- where I is the indicator function. Then Y has a distribution which is a special case of the multinomial distribution with parameter
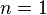 . The sum of
. The sum of 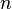 independent and identically distributed such random variables Y constructed from a categorical distribution with parameter
independent and identically distributed such random variables Y constructed from a categorical distribution with parameter 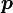 is multinomially distributed with parameters
is multinomially distributed with parameters 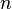 and
and 
- The conjugate prior distribution of a categorical distribution is a Dirichlet distribution.[1] See the section below for more discussion.
- The sufficient statistic from n independent observations is the set of counts (or, equivalently, proportion) of observations in each category, where the total number of trials (=n) is fixed.
- The indicator function of an observation having a value i, equivalent to the Iverson bracket function
![[x=i]](../I/m/2bd4b55c9539155fb4b0da36be8424a1.png) or the Kronecker delta function
or the Kronecker delta function  is Bernoulli distributed with parameter
is Bernoulli distributed with parameter 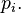
With a conjugate prior
In Bayesian statistics, the Dirichlet distribution is the conjugate prior distribution of the categorical distribution (and also the multinomial distribution). This means that in a model consisting of a data point having a categorical distribution with unknown parameter vector p, and (in standard Bayesian style) we choose to treat this parameter as a random variable and give it a prior distribution defined using a Dirichlet distribution, then the posterior distribution of the parameter, after incorporating the knowledge gained from the observed data, is also a Dirichlet. Intuitively, in such a case, starting from what we know about the parameter prior to observing the data point, we then can update our knowledge based on the data point and end up with a new distribution of the same form as the old one. This means that we can successively update our knowledge of a parameter by incorporating new observations one at a time, without running into mathematical difficulties.
Formally, this can be expressed as follows. Given a model
then the following holds:[1]
This relationship is used in Bayesian statistics to estimate the underlying parameter p of a categorical distribution given a collection of N samples. Intuitively, we can view the hyperprior vector α as pseudocounts, i.e. as representing the number of observations in each category that we have already seen. Then we simply add in the counts for all the new observations (the vector c) in order to derive the posterior distribution.
Further intuition comes from the expected value of the posterior distribution (see the article on the Dirichlet distribution):
This says that the expected probability of seeing a category i among the various discrete distributions generated by the posterior distribution is simply equal to the proportion of occurrences of that category actually seen in the data, including the pseudocounts in the prior distribution. This makes a great deal of intuitive sense: If, for example, there are three possible categories, and we saw category 1 in our observed data 40% of the time, we would expect on average to see category 1 40% of the time in the posterior distribution as well.
(Note that this intuition is ignoring the effect of the prior distribution. Furthermore, it's important to keep in mind that the posterior is a distribution over distributions. Remember that the posterior distribution in general tells us what we know about the parameter in question, and in this case the parameter itself is a discrete probability distribution, i.e. the actual categorical distribution that generated our data. For example, if we saw the 3 categories in the ratio 40:5:55 in our observed data, then ignoring the effect of the prior distribution, we would expect the true parameter — i.e. the true, underlying distribution that generated our observed data — to have the average value of (0.40,0.05,0.55), which is indeed what the posterior tells us. However, the true distribution might actually be (0.35,0.07,0.58) or (0.42,0.04,0.54) or various other nearby possibilities. The amount of uncertainty involved here is specified by the variance of the posterior, which is controlled by the total number of observations – the more data we observe, the less our uncertainty about the true parameter.)
(Technically, the prior parameter 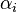 should actually be seen as representing
should actually be seen as representing 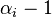 prior observations of category
prior observations of category 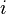 . Then, the updated posterior parameter
. Then, the updated posterior parameter 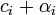 represents
represents 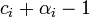 posterior observations. This reflects the fact that a Dirichlet distribution with
posterior observations. This reflects the fact that a Dirichlet distribution with  has a completely flat shape — essentially, a uniform distribution over the simplex of possible values of p. Logically, a flat distribution of this sort represents total ignorance, corresponding to no observations of any sort. However, the mathematical updating of the posterior works fine if we ignore the
has a completely flat shape — essentially, a uniform distribution over the simplex of possible values of p. Logically, a flat distribution of this sort represents total ignorance, corresponding to no observations of any sort. However, the mathematical updating of the posterior works fine if we ignore the 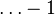 term and simply think of the α vector as directly representing a set of pseudocounts. Furthermore, doing this avoids the issue of interpreting
term and simply think of the α vector as directly representing a set of pseudocounts. Furthermore, doing this avoids the issue of interpreting 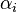 values less than 1.)
values less than 1.)
MAP Estimation
The maximum-a-posteriori estimate of the parameter p in the above model is simply the mode of the posterior Dirichlet distribution, i.e.,[1]
In many practical applications, the only way to guarantee the condition that 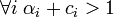 is to set
is to set 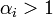 for all i.
for all i.
Marginal likelihood
In the above model, the marginal likelihood of the observations (i.e. the joint distribution of the observations, with the prior parameter marginalized out) is a Dirichlet-multinomial distribution:[1]
This distribution plays an important role in hierarchical Bayesian models, because when doing inference over such models using methods such as Gibbs sampling or variational Bayes, Dirichlet prior distributions are often marginalized out. See the article on this distribution for more details.
Posterior predictive distribution
The posterior predictive distribution of a new observation in the above model is the distribution that a new observation 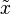 would take given the set
would take given the set 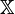 of N categorical observations. As shown in the Dirichlet-multinomial distribution article, it has a very simple form:[1]
of N categorical observations. As shown in the Dirichlet-multinomial distribution article, it has a very simple form:[1]
Note the various relationships among this formula and the previous ones:
- The posterior predictive probability of seeing a particular category is the same as the relative proportion of previous observations in that category (including the pseudo-observations of the prior). This makes logical sense — intuitively, we would expect to see a particular category according to the frequency already observed of that category.
- The posterior predictive probability is the same as the expected value of the posterior distribution. This is explained more below.
- As a result, this formula can be expressed as simply "the posterior predictive probability of seeing a category is proportional to the total observed count of that category", or as "the expected count of a category is the same as the total observed count of the category", where "observed count" is taken to include the pseudo-observations of the prior.
The reason for the equivalence between posterior predictive probability and the expected value of the posterior distribution of p is evident once we re-examine the above formula. As explained in the posterior predictive distribution article, the formula for the posterior predictive probability has the form of an expected value taken with respect to the posterior distribution:
The crucial line above is the third. The second follows directly from the definition of expected value. The third line is particular to the categorical distribution, and follows from the fact that, in the categorical distribution specifically, the expected value of seeing a particular value i is directly specified by the associated parameter pi. The fourth line is simply a rewriting of the third in a different notation, using the notation farther up for an expectation taken with respect to the posterior distribution of the parameters.
Note also what happens in a scenario in which we observe data points one by one and each time consider their predictive probability before observing the data point and updating the posterior. For any given data point, the probability of that point assuming a given category depends on the number of data points already in that category. If a category has a high frequency of occurrence, then new data points are more likely to join that category — further enriching the same category. This type of scenario is often termed a preferential attachment (or "rich get richer") model. This models many real-world processes, and in such cases the choices made by the first few data points have an outsize influence on the rest of the data points.
Posterior conditional distribution
In Gibbs sampling, we typically need to draw from conditional distributions in multi-variable Bayes networks where each variable is conditioned on all the others. In networks that include categorical variables with Dirichlet priors (e.g. mixture models and models including mixture components), the Dirichlet distributions are often "collapsed out" (marginalized out) of the network, which introduces dependencies among the various categorical nodes dependent on a given prior (specifically, their joint distribution is a Dirichlet-multinomial distribution). One of the reasons for doing this is that in such a case, the distribution of one categorical node given the others is exactly the posterior predictive distribution of the remaining nodes.
That is, for a set of nodes 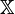 , if we denote the node in question as
, if we denote the node in question as 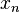 and the remainder as
and the remainder as 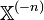 , then
, then
where 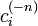 is the number of nodes having category i among the nodes other than node n.
is the number of nodes having category i among the nodes other than node n.
Sampling
The most common way to sample from a categorical distribution uses a type of inverse transform sampling:
Assume we are given a distribution expressed as "proportional to" some expression, with unknown normalizing constant. Then, before taking any samples, we prepare some values as follows:
- Compute the unnormalized value of the distribution for each category.
- Sum them up and divide each value by this sum, in order to normalize them.
- Impose some sort of order on the categories (e.g. by an index that runs from 1 to k, where k is the number of categories).
- Convert the values to a cumulative distribution function (CDF) by replacing each value with the sum of all of the previous values. This can be done in time O(k). The resulting value for the first category will be 0.
Then, each time it is necessary to sample a value:
- Pick a uniformly distributed number between 0 and 1.
- Locate the greatest number in the CDF whose value is less than or equal to the number just chosen. This can be done in time O(log(k)), by binary search.
- Return the category corresponding to this CDF value.
If it is necessary to draw many values from the same categorical distribution, the following approach is more efficient. It draws n samples in O(n) time (assuming an O(1) approximation is used to draw values from the binomial distribution[5]).
function draw_categorical(n) // where n is the number of samples to draw from the categorical distribution
r = 1
s = 0
for i from 1 to k // where k is the number of categories
v = draw from a binomial(n, p[i] / r) distribution // where p[i] is the probability of category i
for j from 1 to v
z[s++] = i // where z is an array in which the results are stored
n = n - v
r = r - p[i]
shuffle (randomly re-order) the elements in z
return z
Sampling via the Gumbel distribution
In machine learning it is typical to parametrize the categorical distribution, 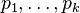 via an unconstrained representation in
via an unconstrained representation in 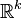 , whose components are given by:
, whose components are given by:
where 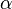 is any real constant. Given this representation,
is any real constant. Given this representation, 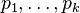 can be recovered using the softmax function, which can then be sampled using the techniques described above. There is however a more direct sampling method that uses samples from the Gumbel distribution.[6] Let
can be recovered using the softmax function, which can then be sampled using the techniques described above. There is however a more direct sampling method that uses samples from the Gumbel distribution.[6] Let 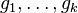 be k independent draws from the standard Gumbel distribution, then
be k independent draws from the standard Gumbel distribution, then
will be a sample from the desired categorical distribution. (If 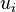 is a sample from the standard uniform distribution, then
is a sample from the standard uniform distribution, then 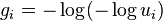 is a sample from the standard Gumbel distribution.)
is a sample from the standard Gumbel distribution.)
See also
- Categorical data
- Categorical variable
Related distributions
- Dirichlet distribution
- Multinomial distribution
- Bernoulli distribution
- Dirichlet-multinomial distribution
Notes
- ↑ However, Bishop does not explicitly use the term categorical distribution.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Minka, T. (2003) Bayesian inference, entropy and the multinomial distribution. Technical report Microsoft Research.
- ↑ Minka, T. (2003), op. cit. Minka uses the Kronecker delta function, similar to but less general than the Iverson bracket.
- ↑ 3.0 3.1 Bishop, C. (2006) Pattern Recognition and Machine Learning, Springer. ISBN 0-387-31073-8
- ↑ Johnson, N.L., Kotz, S., Balakrishnan, N. (1997) Discrete Multivariate Distributions, Wiley. ISBN 0-471-12844-9 (p.105)
- ↑ Agresti, A., An Introduction to Categorical Data Analysis, Wiley-Interscience, 2007, ISBN 978-0-471-22618-5, pp. 25
- ↑ Adams, Ryan. "The Gumbel-Max Trick for Discrete Distributions".
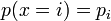
![p(x) = p_1^{[x=1]} \cdots p_k^{[x=k]}](../I/m/cc10881256777a214b3da27a751d9869.png)
![p(x) = [x=1]\cdot p_1 \, + \dots + \, [x=k]\cdot p_k](../I/m/e08c8bd9d89d21127d3ea441f871e232.png)

![f(x| \boldsymbol{p} ) = \prod_{i=1}^k p_i^{[x=i]} ,](../I/m/e41d5d12d0c09c589c0b5d3eea6bb137.png)
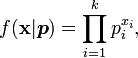
![\mathbb{E} \left[ \mathbf{x} \right] = \boldsymbol{p}](../I/m/a611709c883c977127d0ffec8f55b55f.png)
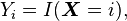
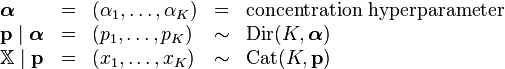
![\begin{array}{lclcl}
\mathbf{c} &=& (c_1, \ldots, c_K) &=& \text{number of occurrences of category }i = \sum_{j=1}^N [x_j=i] \\
\mathbf{p} \mid \mathbb{X},\boldsymbol\alpha &\sim& \operatorname{Dir}(K,\mathbf{c}+\boldsymbol\alpha) &=& \operatorname{Dir}(K,c_1+\alpha_1,\ldots,c_K+\alpha_K)
\end{array}](../I/m/e0104d3723147d5364a4dc58c3b7c637.png)
![\mathbb{E}[p_i \mid \mathbb{X},\boldsymbol\alpha] = \frac{c_i+\alpha_i}{N+\sum_k\alpha_k}](../I/m/802ee560c4411fe7d5364977fa89094f.png)
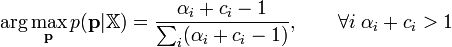
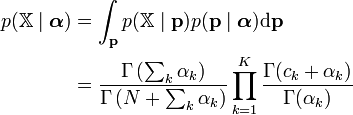
![\begin{align}
p(\tilde{x}=i\mid\mathbb{X},\boldsymbol{\alpha}) &= \int_{\mathbf{p}}p(\tilde{x}=i\mid\mathbf{p})\,p(\mathbf{p}\mid\mathbb{X},\boldsymbol{\alpha})\,\textrm{d}\mathbf{p} \\
&=\, \frac{c_i + \alpha_i}{N+\sum_k \alpha_k} \\
&=\, \mathbb{E}[p_i \mid \mathbb{X},\boldsymbol\alpha] \\
&\propto\, c_i + \alpha_i. \\
\end{align}](../I/m/c24b1a075dbbf66e6062c969651aee8a.png)
![\begin{align}
p(\tilde{x}=i\mid\mathbb{X},\boldsymbol{\alpha}) &= \int_{\mathbf{p}}p(\tilde{x}=i\mid\mathbf{p})\,p(\mathbf{p}\mid\mathbb{X},\boldsymbol{\alpha})\,\textrm{d}\mathbf{p} \\
&=\, \mathbb{E}_{\mathbf{p}\mid\mathbb{X},\boldsymbol{\alpha}} \left[p(\tilde{x}=i\mid\mathbf{p})\right] \\
&=\, \mathbb{E}_{\mathbf{p}\mid\mathbb{X},\boldsymbol{\alpha}} \left[p_i\right] \\
&=\, \mathbb{E}[p_i \mid \mathbb{X},\boldsymbol\alpha]. \\
\end{align}](../I/m/b381b9e1e7c555f72b3254d5c55a1272.png)