Carboxysome

Carboxysomes are bacterial microcompartments that contain enzymes involved in carbon fixation.[1] Carboxysomes are made of polyhedral protein shells about 80 to 140 nanometres in diameter. These compartments are thought to concentrate carbon dioxide to overcome the inefficiency of RuBisCO - the predominant enzyme in carbon fixation and the rate limiting enzyme in the Calvin cycle. These organelles are found in all cyanobacteria and many chemotrophic bacteria that fix carbon dioxide.[2]
Carboxysomes are an example of a wider group of protein micro-compartments that have dissimilar functions, but similar structures, based on homology of the two shell protein families.[3]
Discovery
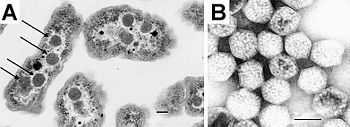
Using electron microscopy the first carboxysomes were seen in 1956, in the cyanobacterium Phormidium uncinatum[2][5] in the early 1960s similar polyhedral objects were observed in other cyanobacteria.[6] These structures were named polyhedral bodies in 1961 and over the next few years were also discovered in some chemotrophic bacteria that fixed carbon dioxide (for example, Halothiobacillus, Acidithiobacillus, Nitrobacter and Nitrococcus).[2][7]
Carboxysomes were first purified from Thiobacillus neapolitanus in 1973 and shown to contain RuBisCo, held within a rigid outer covering.[8] The authors proposed that since these appeared to be organelles involved in carbon fixation, they should be called carboxysomes.[9]
Architecture

]

Structurally, carboxysomes are icosahedral, or quasi-icosahedral, typically around 80 to 120 nm in diameter.[10] The carboxysome has an outer shell composed of a few thousand protein subunits, which encapsulates the two carbon fixing enzymes, carbonic anhydrase and RuBisCO. Proteins known to form the shell have been structurally characterized by X-ray crystallography.[11] The protein that constitutes the majority of the shell forms a cyclical hexamer. These hexamers constitute the basic building blocks of the shell.[11] X-ray structural data suggest that the hexamers assemble further in a side-by-side fashion to form a tightly packed molecular layer, which represents the outer shell. Small pores are present through the hexamers and may serve as the route for diffusion of substrates (bicarbonate and ribulose-1,5-bisphosphate) and products (3-phosphoglycerate) into and out of the carboxysome. Positively charged amino acids in the pores might help promote the diffusion of the negatively charged substrates and products. Other minor structural components of the shell that have been characterized include pentameric proteins, which have been proposed to occupy the vertices of the icosahedral shell.[10]
A number of viral capsids are also constructed from hexameric and pentameric proteins, but whether any evolutionary relationship exists between the carboxysome and viral capsids is unknown. Electron cryo-tomography studies have confirmed the approximately icosahedral geometry of the carboxysome, and have visualized enzymes molecules inside, presumed to be RuBisCO, arranged in a few concentric layers.[12][13] In addition, non-icosahedral faceted shapes of some carboxysomes can naturally be explained within the elastic theory of heterogeneous thin shells. [14]
Studies in Halothiobacillus neapolitanus have shown that empty shells of normal shape and composition are assembled in carboxysomal RuBisCO-lacking mutants suggesting that carboxysome shell biogenesis and enzyme sequestration are two independent, but functionally linked processes. Intriguingly, carboxysomes of Halothiobacillus neapolitanus have been found to accommodate chimeric and heterologous species of RuBisCO and it is the large subunit of RuBisCO that determines whether the enzyme is sequestered into carboxysomes or not.[15]
Occurrence
Carboxysomes are found in all cyanobacteria, some nitrifying bacteria, and thiobacilli.[2]
References
- ↑ Badger MR, Price GD (February 2003). "CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution". J. Exp. Bot. 54 (383): 609–22. doi:10.1093/jxb/erg076. PMID 12554704.
- ↑ 2.0 2.1 2.2 2.3 Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (August 2008). "Protein-based organelles in bacteria: carboxysomes and related microcompartments". Nat. Rev. Microbiol. 6 (9): 681–691. doi:10.1038/nrmicro1913. PMID 18679172.
- ↑ Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM (2001). "Microcompartments in Prokaryotes: Carboxysomes and Related Polyhedra". Appl. Environ. Microbiol. 67 (12): 5351–61. doi:10.1128/AEM.67.12.5351-5361.2001. PMC 93316. PMID 11722879.
- ↑ Tsai Y, Sawaya MR, Cannon GC et al. (June 2007). "Structural Analysis of CsoS1A and the Protein Shell of the Halothiobacillus neapolitanus Carboxysome". PLoS Biol. 5 (6): e144. doi:10.1371/journal.pbio.0050144. PMC 1872035. PMID 17518518.
- ↑ Drews G, Niklowitz W (1956). "[Cytology of Cyanophycea. II. Centroplasm and granular inclusions of Phormidium uncinatum.]". Arch Mikrobiol (in German) 24 (2): 147–62. PMID 13327992.
- ↑ Gantt E, Conti SF (1 March 1969). "Ultrastructure of Blue-Green Algae". J. Bacteriol. 97 (3): 1486–93. PMC 249872. PMID 5776533.
- ↑ Shively JM (1974). "Inclusion bodies of prokaryotes". Annu. Rev. Microbiol. 28 (0): 167–87. doi:10.1146/annurev.mi.28.100174.001123. PMID 4372937.
- ↑ Shively JM, Ball F, Brown DH, Saunders RE (November 1973). "Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus". Science 182 (112): 584–6. doi:10.1126/science.182.4112.584. PMID 4355679.
- ↑ Shively JM, Ball FL, Kline BW (1 December 1973). "Electron Microscopy of the Carboxysomes (Polyhedral Bodies) of Thiobacillus neapolitanus". J. Bacteriol. 116 (3): 1405–11. PMC 246500. PMID 4127632.
- ↑ 10.0 10.1 Tanaka S, Kerfeld CA, Sawaya MR et al. (February 2008). "Atomic-level models of the bacterial carboxysome shell". Science 319 (5866): 1083–6. doi:10.1126/science.1151458. PMID 18292340.
- ↑ 11.0 11.1 Kerfeld, CA, Sawaya, MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (2005). "Protein structures forming the shell of primitive bacterial organelles". Science 309 (5736): 936–8. doi:10.1126/science.1113397. PMID 16081736.
- ↑ Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM (2006). "Structure of Halothiobacillus neapolitanus Carboxysomes by Cryo-Electron Tomography". J. Mol. Biol. 364 (3): 526–35. doi:10.1016/j.jmb.2006.09.024. PMC 1839851. PMID 17028023.
- ↑ Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ (2007). "The Structure of Isolated Synechococcus Strain WH8102 Carboxysomes as Revealed by Electron Cryotomography". J. Mol. Biol. 372 (3): 764–73. doi:10.1016/j.jmb.2007.06.059. PMC 2453779. PMID 17669419.
- ↑ Vernizzi G, Sknepnek R, Olvera de la Cruz M (2011). "Platonic and Archimedean geometries in multicomponent elastic membranes". Proc. Natl. Acad. Sci. USA 108 (11): 4292–4296. doi:10.1073/pnas.1012872108. PMC 3060260. PMID 21368184.
- ↑ Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC (October 2008). Rutherford, Julian, ed. "Halothiobacillus neapolitanus Carboxysomes Sequester Heterologous and Chimeric RubisCO Species". PLoS ONE 3 (10): e3570. doi:10.1371/journal.pone.0003570. PMC 2570492. PMID 18974784.