CYP27A1
CYP27A1 is a gene encoding a cytochrome P450 oxidase, and is commonly known as sterol 27-hydroxylase. This enzyme is located in many different tissues where it is found within the mitochondria. It is most prominently involved in the biosynthesis of bile acids.
Function
CYP27A1 participates in the degradation of cholesterol to bile acids in both the classic and acidic pathways.[1] It is the initiating enzyme in the acidic pathway to bile acids, yielding oxysterols by introducing a hydroxyl group to the carbon at the 27 position in cholesterol. In the acidic pathway, it produces 27-hydroxycholesterol from cholesterol whereas in the classic or neutral pathway, it produces 3β-hydroxy-5-cholestenoic acid.
It is also involved in the metabolism of vitamin D3.[2]
While CYP27A1 is present in many different tissues, its function in these tissues is largely uncharacterized. In macrophages, 27-hydroxycholesterol generated by this enzyme may be helpful against the production of inflammatory factors associated with cardiovascular disease.[3]
Clinical significance
Mutations in CYP27A1 are associated with cerebrotendineous xanthomatosis, a rare lipid storage disease.
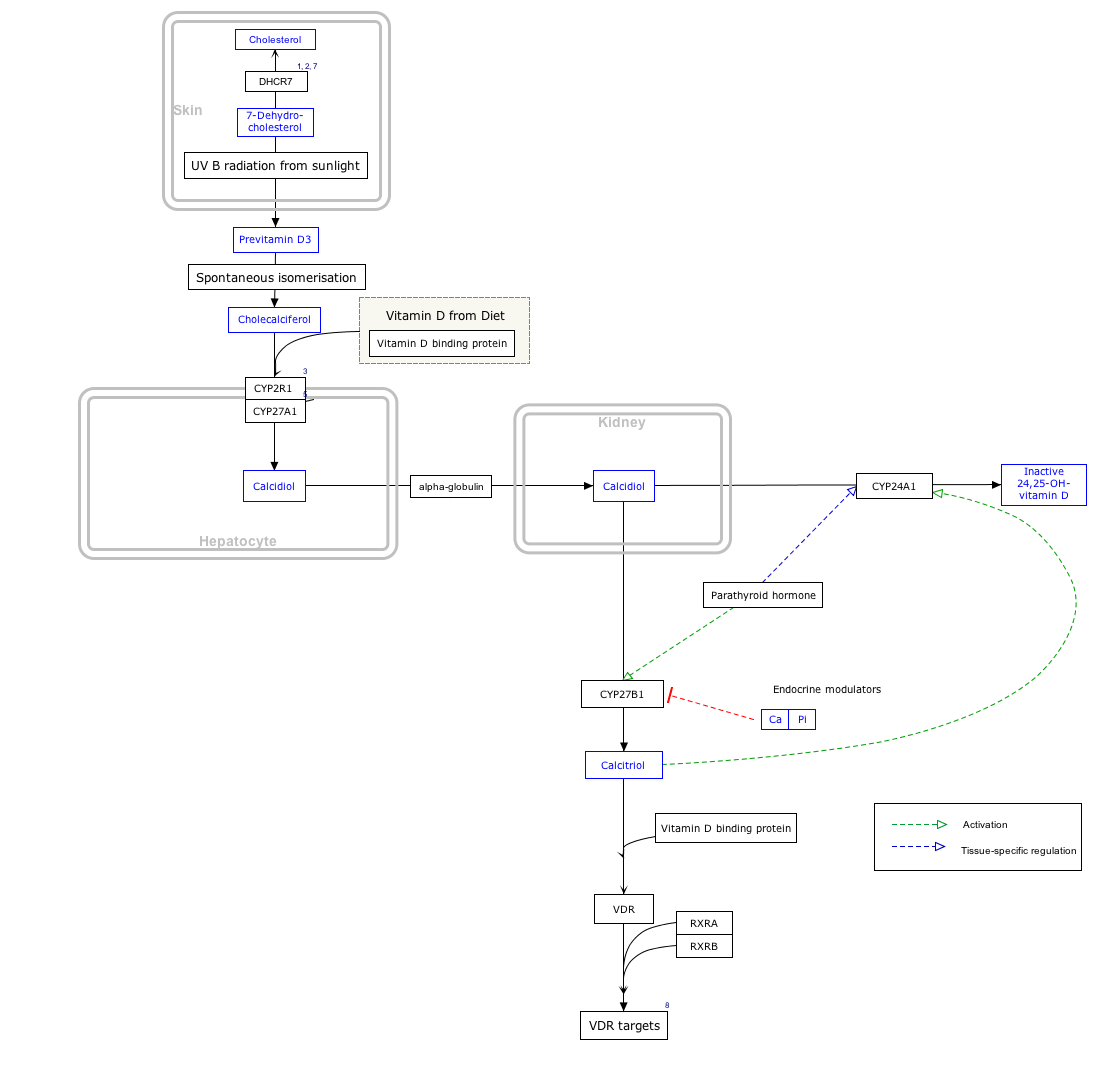
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
Vitamin D Synthesis Pathway edit
- ↑ The interactive pathway map can be edited at WikiPathways: "VitaminDSynthesis_WP1531".
References
- ↑ Chiang JY (February 1998). "Regulation of bile acid synthesis". Front Biosci 3: d176–193. PMID 9450986.
- ↑ Sakaki T, Kagawa N, Yamamoto K, Inouye K (January 2005). "Metabolism of vitamin D3 by cytochromes P450". Front. Biosci. 10: 119–34. PMID 15574355.
- ↑ Taylor JM, Borthwick F, Bartholomew C, Graham A (June 2010). "Overexpression of steroidogenic acute regulatory protein increases macrophage cholesterol efflux to apolipoprotein AI". Cardiovasc Res 86 (3): 526–534. doi:10.1093/cvr/cvq015. PMID 20083572.
Further reading
- Cali JJ, Russell DW (1991). "Characterization of human sterol 27-hydroxylase. A mitochondrial cytochrome P-450 that catalyzes multiple oxidation reaction in bile acid biosynthesis.". J. Biol. Chem. 266 (12): 7774–8. PMID 1708392.
- Cali JJ, Hsieh CL, Francke U, Russell DW (1991). "Mutations in the bile acid biosynthetic enzyme sterol 27-hydroxylase underlie cerebrotendinous xanthomatosis.". J. Biol. Chem. 266 (12): 7779–83. PMID 2019602.
- Guo YD, Strugnell S, Back DW, Jones G (1993). "Transfected human liver cytochrome P-450 hydroxylates vitamin D analogs at different side-chain positions.". Proc. Natl. Acad. Sci. U.S.A. 90 (18): 8668–72. doi:10.1073/pnas.90.18.8668. PMC 47419. PMID 7690968.
- Kim KS; Kubota S; Kuriyama M et al. (1994). "Identification of new mutations in sterol 27-hydroxylase gene in Japanese patients with cerebrotendinous xanthomatosis (CTX)". J. Lipid Res. 35 (6): 1031–9. PMID 7915755.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Leitersdorf E; Reshef A; Meiner V et al. (1993). "Frameshift and splice-junction mutations in the sterol 27-hydroxylase gene cause cerebrotendinous xanthomatosis in Jews or Moroccan origin". J. Clin. Invest. 91 (6): 2488–96. doi:10.1172/JCI116484. PMC 443309. PMID 8514861.
- Chen W; Kubota S; Kim KS et al. (1997). "Novel homozygous and compound heterozygous mutations of sterol 27-hydroxylase gene (CYP27) cause cerebrotendinous xanthomatosis in three Japanese patients from two unrelated families". J. Lipid Res. 38 (5): 870–9. PMID 9186905.
- Reiss AB; Martin KO; Rojer DE et al. (1997). "Sterol 27-hydroxylase: expression in human arterial endothelium". J. Lipid Res. 38 (6): 1254–60. PMID 9215552.
- Suzuki Y; Yoshitomo-Nakagawa K; Maruyama K et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Chen W; Kubota S; Ujike H et al. (1998). "A novel Arg362Ser mutation in the sterol 27-hydroxylase gene (CYP27): its effects on pre-mRNA splicing and enzyme activity". Biochemistry 37 (43): 15050–6. doi:10.1021/bi9807660. PMID 9790667.
- Shiga K; Fukuyama R; Kimura S et al. (1999). "Mutation of the sterol 27-hydroxylase gene (CYP27) results in truncation of mRNA expressed in leucocytes in a Japanese family with cerebrotendinous xanthomatosis". J. Neurol. Neurosurg. Psychiatr. 67 (5): 675–7. doi:10.1136/jnnp.67.5.675. PMC 1736608. PMID 10519880.
- Gascon-Barré M; Demers C; Ghrab O et al. (2001). "Expression of CYP27A, a gene encoding a vitamin D-25 hydroxylase in human liver and kidney". Clin. Endocrinol. (Oxf) 54 (1): 107–15. doi:10.1046/j.1365-2265.2001.01160.x. PMID 11167933.
- Johnston TP, Nguyen LB, Chu WA, Shefer S (2001). "Potency of select statin drugs in a new mouse model of hyperlipidemia and atherosclerosis". International journal of pharmaceutics 229 (1–2): 75–86. doi:10.1016/S0378-5173(01)00834-1. PMID 11604260.
- Toba H; Fukuyama R; Sasaki M et al. (2002). "A Japanese patient with cerebrotendinous xanthomatosis has different mutations within two functional domains of CYP27". Clin. Genet. 61 (1): 77–8. doi:10.1034/j.1399-0004.2002.610116.x. PMID 11903362.
- Lamon-Fava S; Schaefer EJ; Garuti R et al. (2002). "Two novel mutations in the sterol 27-hydroxylase gene causing cerebrotendinous xanthomatosis". Clin. Genet. 61 (3): 185–91. doi:10.1034/j.1399-0004.2002.610303.x. PMID 12000359.
- Björkhem I; Araya Z; Rudling M et al. (2002). "Differences in the regulation of the classical and the alternative pathway for bile acid synthesis in human liver. No coordinate regulation of CYP7A1 and CYP27A1". J. Biol. Chem. 277 (30): 26804–7. doi:10.1074/jbc.M202343200. PMID 12011083.
- von Bahr S; Movin T; Papadogiannakis N et al. (2002). "Mechanism of accumulation of cholesterol and cholestanol in tendons and the role of sterol 27-hydroxylase (CYP27A1)". Arterioscler. Thromb. Vasc. Biol. 22 (7): 1129–35. doi:10.1161/01.ATV.0000022600.61391.A5. PMID 12117727.
- Meir K; Kitsberg D; Alkalay I et al. (2002). "Human sterol 27-hydroxylase (CYP27) overexpressor transgenic mouse model. Evidence against 27-hydroxycholesterol as a critical regulator of cholesterol homeostasis". J. Biol. Chem. 277 (37): 34036–41. doi:10.1074/jbc.M201122200. PMID 12119285.
- Lee MJ; Huang YC; Sweeney MG et al. (2002). "Mutation of the sterol 27-hydroxylase gene ( CYP27A1) in a Taiwanese family with cerebrotendinous xanthomatosis". J. Neurol. 249 (9): 1311–2. doi:10.1007/s00415-002-0762-9. PMID 12242561.
- Strausberg RL; Feingold EA; Grouse LH et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
External links
- GeneReviews/NCBI/NIH/UW entry on Cerebrotendinous Xanthomatosis
- Cytochrome P-450 CYP27A1 at the US National Library of Medicine Medical Subject Headings (MeSH)
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||