BioBrick
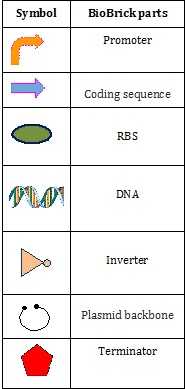
BioBrick parts are DNA sequences which conform to a restriction-enzyme assembly standard.[1][2] These Lego-like building blocks are used to design and assemble synthetic biological circuits, which would then be incorporated into living cells such as Escherichia coli cells to construct new biological systems.[3] Examples of BioBrick parts include promoters, ribosomal binding sites (RBS), coding sequences and terminators.
Parts, device and system
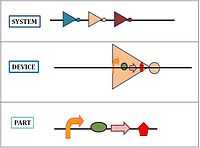
The BioBrick parts are used by applying engineering principles of abstraction and modularization. BioBrick parts form the base of the hierarchical system on which synthetic biology is based. There are three levels to the hierarchy: parts, device and system.
- Parts: Pieces of DNA that form a functional unit (for example promoter, RBS, etc.)
- Device: Collection set of parts with defined function. In simple terms, a set of complementary BioBrick parts put together forms a device.
- System: Combination of a set of devices that performs high-level tasks.
History
The first attempt to create a list of standard biological parts was in 1996, by Rebatchouk et al. This team introduced a cloning strategy for the assembly of short DNA fragments. However, this early attempt was not widely recognised by the scientific research community at the time.[2][4] In 1999, Arkin and Endy realized that the heterogeneous elements that made up a genetic circuit were lacking standards, so they proposed a list of standard biological parts.[5] BioBricks were described and introduced by Tom Knight at MIT in 2003. Since then, various research groups have utilized and used the BioBrick standard parts to engineer novel biological devices and systems.
BioBrick Assembly standard
The BioBrick assembly standard was introduced to overcome the lack of standardization posed by traditional molecular cloning methods. The BioBrick assembly standard is a more reliable approach for combining parts to form larger composites. The assembly standard enables two groups of synthetic biologists in different parts of the world to re-use a BioBrick part without going through the whole cycle of design and manipulation.[2] This means the newly designed part can be used by other teams of researchers more easily. Besides that, when compared to the old fashioned ad hoc cloning method, the assembly standard process is faster and promotes automation.[6] The BioBrick assembly standard 10 was the first assembly standard to be introduced. Over the years, several other assembly standards, such as the Biofusion standard and Freiburg standard have been developed.
BioBrick assembly standard 10
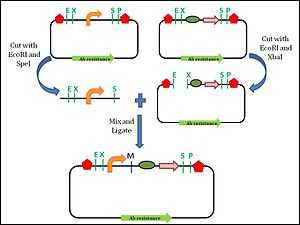
Assembly standard 10 was developed by Tom Knight, and is the most widely used assembly standard. It involves the use of restriction enzymes. Every BioBrick part is a DNA sequence which is carried by a circular plasmid, which acts a vector.[7] The vector acts as a transport system to carry the BioBrick parts. The first approach towards a BioBrick standard was the introduction of standard sequences, the prefix and suffix sequences, which flank the 5′ and 3′ ends of the DNA part respectively.[8] These standard sequences encode specific restriction enzyme sites. The prefix sequence encodes EcoRI (E) and Xbal (X) sites, while the suffix sequence encodes SpeI (S) and PstI (P) sites. The prefix and the suffix are not considered part of the BioBrick part.[3] To facilitate the assembly process, the BioBrick part itself must not contain any of these restriction sites. During the assembly of two different parts, one of the plasmids is digested with SpeI and Xbal. The plasmid carrying the other BioBrick part is digested with EcoRI and Xbal. This leaves both plasmids with 4 base pair (bp) overhangs at the 5’ and 3’ ends. The EcoRI sites will ligate since they are complementary to each other. The Xbal and SpeI sites will also ligate as the digestion produces compatible ends. Now, both the DNA parts are in one plasmid. The ligation produces an 8 base pair ‘scar’ site between the two BioBrick parts. Since the scar site is a hybrid of the Xbal and SpeI sites, it is not recognized by either restriction enzyme.[8] The prefix and suffix sequences remain unchanged by this digestion and ligation process, which allows for subsequent assembly steps with more BioBrick parts.
This assembly is an idempotent process: multiple applications don’t change the end product, and maintain the prefix and suffix. Although the BioBrick standard assembly allows for the formation of functional modules, there is a limitation to this standard 10 approach. The 8 bp scar site does not allow the creation of a fusion protein.[7] The scar site causes a frame shift which prevents the continuous reading of codons, which is required for the formation of fusion protein.
Silver (Biofusion) standard
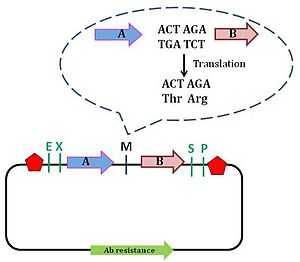
Pam Silver’s lab created the Silver assembly standard to overcome the issue surrounding the formation of fusion protein. This assembly standard is also known as Biofusion standard, and is an improvement of the BioBrick assembly standard 10. Silver’s standard involves deletion of one nucleotide from the Xbal and SpeI site, which shortens the scar site by 2 nucleotides, which now forms a 6 bp scar sequence. The 6 bp sequence allows the reading frame to be maintained.The scar sequence codes for the amino acid threonine (ACT) and arginine (AGA).[9] This minor improvement allows for the formation of in-frame fusion protein. However, arginine's being a large, charged amino acid is a disadvantage to the Biofusion assembly technique: these properties of arginine result in the destabilisation of the protein by the N-end rule.
Freiburg standard
The 2007 Freiburg iGEM team introduced a new assembly standard to overcome the disadvantages of the existing Biofusion standard technique. The Freiburg team created a new set of prefix and suffix sequences by introducing additional restriction enzyme sites, AgeI and NgoMIV to the existing prefix and suffix respectively. These newly introduced restriction enzyme sites are BioBrick standard compatible. The Freiburg standard still forms a 6 bp scar site, but the scar sequence (ACCGGC) now codes for threonine and glycine respectively. This scar sequence results in a much more stable protein[10] as the glycine forms a stable N-terminal, unlike the arginine, which signals for N-terminal degradation. The assembly technique proposed by the Freiburg team diminishes the limitations of the Biofusion standard.
Assembly method
Different methods are used when it comes to assembling BioBricks. This is because, some standards require different materials and methods (use of different restriction enzymes), while others are due to preferences in protocol because some methods of assembly have higher efficiency and is user-friendly.
3 Antibiotic (3A) assembly
The 3A assembly method is the most commonly used, as its compatible with assembly Standard 10, Silver standard as well as the Freiburg standard. This assembly method involves two BioBrick parts and a destination plasmid. The destination plasmid contains the toxic(lethal) gene, to ease the selection of correctly assembled plasmid. The destination plasmid also have a different antibiotic resistance genes than the plasmids carrying the BioBrick parts. All three plasmids are digested with appropriate restriction enzyme and then allowed to ligate. Only the correctly assembled part will produce a viable composite part contained in the destination plasmid. This allows a good selection as only the correctly assembled BioBrick parts survives.
Parts Registry
The MIT group lead by Tom Knight that developed BioBricks and International Genetically Engineered Machines (iGEM) competition are also the pioneers of The Registry of Standard Biological Parts (Registry).[11] Registry being one of the foundations of synthetic biology, provides web-based information and data on over 2000 BioBrick parts. The Registry contains:
- Information and characterisation data for all parts, device and system
- Includes a catalogue which describes the function, performance and design of each part
Every BioBrick part has its unique identification code which makes the search for the desired BioBrick part easier (for example, BBa_J23100, a constitutive promoter).[2] The registry is open access, whereby anyone can submit a BioBrick part. Most of the BioBrick submission is from students participating in the annual iGEM competition hosted every summer. The Registry allows exchange of data and materials online which allows rapid re-use and modifications of parts by the participating community.
Professional parts registries have also been developed. Since most of the BioBrick parts are submitted by undergraduates as part of the iGEM competition, the parts may lack important characterisation data and metadata which would be essential when it comes to designing and modelling the functional components.[11] One example of a professional parts registry is the USA-based publically-funded facility, The International Open Facility Advancing Biotechnology (BIOFAB), which contains detailed descriptions of each biological part. It is also an open-source registry, and is available commercially. BIOFAB aims to catalogue high-quality BioBrick parts to accommodate the needs of professional synthetic biology community.
The BioBrick Foundation (BBF) is an public-benefit organisation established to promote the use of standardized BioBrick parts on a scale beyond the iGEM competition. The BBF is currently working on the derivation of standard framework to promote the production high quality BioBrick parts which would be freely available to everyone.[12]
See also
References
- ↑ "Tom Knight (2003). Idempotent Vector Design for Standard Assembly of Biobricks". Retrieved 2014-09-26.
- ↑ 2.0 2.1 2.2 2.3 Knight, Thomas F; Reshma P Shetty; Drew Endy (14 April 2008). "Engineering BioBrick vectors from BioBrick parts". Journal of Biological Engineering 2 (5): 1–12. doi:10.1186/1754-1611-2-5. PMC 2373286. PMID 18410688. Retrieved 27 March 2014.
- ↑ 3.0 3.1 "SynBio Standards -BioBrick" (PDF). Retrieved 27 March 2014.
- ↑ Rebatchouk, Dmitri; Daraselia, N.; Narita, J. O. (1 October 1996). "NOMAD: a versatile strategy for in vitro DNA manipulation applied to promoter analysis and vector design.". Proceedings of the National Academy of Sciences 93 (20): 10891–10896. doi:10.1073/pnas.93.20.10891. Retrieved 27 March 2014.
- ↑ Arkin, Adam. "A Standard Parts List for Biological Circuitry" (PDF). Retrieved 27 March 2014.
- ↑ "j5 automated DNA assembly-The BioBrick approach".
- ↑ 7.0 7.1 Sleight, S. C.; Bartley, B. A.; Lieviant, J. A.; Sauro, H. M. (12 April 2010). "In-Fusion BioBrick assembly and re-engineering". Nucleic Acids Research 38 (8): 2624–2636. doi:10.1093/nar/gkq179.
- ↑ 8.0 8.1 Knight, Thomas F.; Reshma Shetty; Meagan Lizarazo; Randy Rettberg (2011). "Chapter 13:Assembly of BioBrick Standard Biological Parts Using Three Antibiotic Assembly" (PDF). Elsevier 498: 311–325. doi:10.1016/B978-0-12-385120-8.00013-9. Retrieved 27 March 2014.
- ↑ Silver, Pamela A.; Ira E. Phillips (April 18, 2006). "A New Biobrick Assembly Strategy Designed for Facile Protein Engineering" (PDF). Harvard Medical School: 1–6.
- ↑ Muller, Kristian M. "http://dspace.mit.edu/bitstream/handle/1721.1/45140/BBF_RFC%2025.pdf?sequence=1" (PDF). MIT. Retrieved 27 March 2014.
- ↑ 11.0 11.1 Baldwin, Geoff (2012). Synthetic Biology A Primer. London: Imperial College Pr. ISBN 1848168632.
- ↑ "About BioBricks Foundation". Retrieved 27 March 2014.