Beta-ureidopropionase
| beta-ureidopropionase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 3.5.1.6 | ||||||||
| CAS number | 9027-27-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
In enzymology, a beta-ureidopropionase (EC 3.5.1.6) is an enzyme that catalyzes the chemical reaction
- N-carbamoyl-beta-alanine + H2O
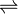 beta-alanine + CO2 + NH3
beta-alanine + CO2 + NH3
Thus, the two substrates of this enzyme are N-carbamoyl-beta-alanine and H2O, whereas its 3 products are beta-alanine, CO2, and NH3.
This enzyme belongs to the family of hydrolases, those acting on carbon-nitrogen bonds other than peptide bonds, specifically in linear amides. The systematic name of this enzyme class is N-carbamoyl-beta-alanine amidohydrolase. This enzyme participates in 3 metabolic pathways: pyrimidine metabolism, beta-alanine metabolism, and pantothenate and coenzyme A biosynthesis.
Structural studies
As of late 2007, 6 structures have been solved for this class of enzymes, with PDB accession codes 1R3N, 1R43, 2V8D, 2V8G, 2V8H, and 2V8V.
References
- CAMPBELL LL (1960). "Reductive degradation of pyrimidines. 5. Enzymatic conversion of N-carbamyl-beta-alanine to beta-alanine, carbon dioxide, and ammonia". J. Biol. Chem. 235: 2375–8. PMID 13849303.
- CARAVACA J, GRISOLIA S (1958). "Enzymatic decarbamylation of carbamyl beta-alanine and carbamyl beta-aminoisobutyric acid". J. Biol. Chem. 231 (1): 357–65. PMID 13538975.
- Traut TW, Loechel S (1984). "Pyrimidine catabolism: individual characterization of the three sequential enzymes with a new assay". Biochemistry. 23 (11): 2533–9. doi:10.1021/bi00306a033. PMID 6433973.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||