Azobenzene reductase
| azobenzene reductase (azoreductase) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.7.1.6 | ||||||||
| CAS number | 9029-31-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
Azobenzene reductase also known as azoreductase (EC 1.7.1.6) is an enzyme that catalyzes the chemical reaction:
- N,N-dimethyl-1,4-phenylenediamine + aniline + NADP+
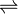 4-(dimethylamino)azobenzene + NADPH + H+
4-(dimethylamino)azobenzene + NADPH + H+
The 3 substrates of this enzyme are N,N-dimethyl-1,4-phenylenediamine, aniline, and nicotinamide adenine dinucleotide phosphate ion, whereas its 3 products are 4-(dimethylamino)azobenzene, nicotinamide adenine dinucleotide phosphate, and hydrogen ion.[1]
This enzyme belongs to the family of oxidoreductases, specifically those acting on other nitrogenous compounds as donors with NAD+ or NADP+ as acceptor.
Mechanism
The reaction catalyzed by this enzyme proceeds via a ping-pong mechanism[2] by using 2 equivalents of NAD(P)H to reduce one equivalent of the azo compound substrate (for example methyl red where Ar = p-dimethylaniline and Ar' = o-benzoic acid) into two equivalents of aniline product:
- Ar–N=N–Ar' + 2(NAD(P)H + H+)
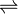 Ar–NH2 + NH2–Ar' + 2NAD(P)+
Ar–NH2 + NH2–Ar' + 2NAD(P)+
Substrate specificity
Most azoreductase isoenzymes can reduce methyl red, but are not able to reduce sulfonated azo dyes. The unique azoreductase isozyme from Bacillus sp. B29 has the ability to reduce sulfonated azo dyes however.[3]
Nomenclature
The systematic name of this enzyme class is N,N-dimethyl-1,4-phenylenediamine, aniline:NADP+ oxidoreductase. Other names in common use include:
- azo reductase,
- azoreductase,
- azo-dye reductase,
- dibromopropylaminophenylazobenzoic azoreductase,
- dimethylaminobenzene reductase,
- methyl red azoreductase,
- N,N-dimethyl-4-phenylazoaniline azoreductase,
- NAD(P)H:1-(4'-sulfophenylazo)-2-naphthol oxidoreductase,
- NADPH2-dependent azoreductase,
- NADPH2:4-(dimethylamino)azobenzene oxidoreductase,
- NC-reductase,
- new coccine (NC)-reductase,
- nicotinamide adenine dinucleotide (phosphate) azoreductase,
- orange I azoreductase,
- orange II azoreductase,
- p-aminoazobenzene reductase, and
- p-dimethylaminoazobenzene azoreductase.
Structural studies
As of late 2007, 3 structures have been solved for this class of enzymes, with PDB accession codes 1NNI, 1V4B, and 2D5I. Please check the last updated data on RCSB PDB site.
References
- ↑ Mueller GC and Miller JA (October 1949). "The reductive cleavage of 4-dimethylaminoazobenzene by rat liver; the intracellular distribution of the enzyme system and its requirement for triphosphopyridine nucleotide". J. Biol. Chem. 180 (3): 1125–36. PMID 18139207.
- ↑ Ooi T, Shibata T, Sato R, Ohno H, Kinoshita S, Thuoc TL, Taguchi S (May 2007). "An azoreductase, aerobic NADH-dependent flavoprotein discovered from Bacillus sp.: functional expression and enzymatic characterization". Appl. Microbiol. Biotechnol. 75 (2): 377–86. doi:10.1007/s00253-006-0836-1. PMID 17546472.
- ↑ Ooi T, Shibata T, Matsumoto K, Kinoshita S, Taguchi S (May 2009). "Comparative enzymatic analysis of azoreductases from Bacillus sp. B29". Biosci. Biotechnol. Biochem. 73 (5): 1209–11. doi:10.1271/bbb.80872. PMID 19420689.
Further reading
- Suzuki Y, Yoda T, Ruhul A, Sugiura W (2001). "Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soil". J. Biol. Chem. 276 (12): 9059–65. doi:10.1074/jbc.M008083200. PMID 11134015.
- Matsumoto K, Mukai Y, Ogata D, Shozui F, Nduko JM, Taguchi S, Ooi T (2009). "Characterization of thermostable FMN-dependent NADH azoreductase from the moderate thermophile Geobacillus stearothermophilus". Appl. Microbiol. Biotechnol. 86 (5): 1431–8. doi:10.1007/s00253-009-2351-7. PMID 19997911.