Archaic human admixture with modern humans
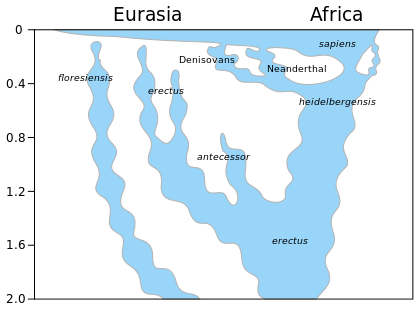
There have been several instances of archaic human admixture with modern humans through interbreeding of modern humans with Neanderthals, Denisovans, and/or possibly other archaic humans over the course of human history. Neanderthal-derived DNA accounts for an estimated 1–4% of the Eurasian genome, but it is significantly absent or uncommon in the genome of most Sub-Saharan African people. In Oceanian and Southeast Asian populations, there's a relative increase of Denisovan-derived DNA. An estimated 4–6% of the Melanesian genome is derived from Denisovans. Recent non-comparative DNA analyses—as there has been no specimens discovered—suggest that African populations have a genetic contribution from a now-extinct archaic African hominin population. Nevertheless, there still are some doubts about the recent admixture event among a number of researchers. Ancient sub-population structure ancestral to modern humans, Neanderthals, Denisovans, and other possible archaic humans has been proposed as an alternative explanation for the observed genetic similarities.
Neanderthals
Genetics
Through whole-genome sequencing, a 2010 draft sequence of the Neanderthal genome revealed that Neanderthals shared more alleles with Eurasian populations (e.g. French, Han Chinese, and Papua New Guinean) than with Sub-Saharan African populations (e.g. Yoruba and San).[2] According to the study, the observed excess of genetic similarity is best explained by recent gene flow from Neanderthals to modern humans after the migration out of Africa.[2] The proportion of Neanderthal-derived ancestry was estimated to be 1–4% of the Eurasian genome.[2] In 2013, the same team of researchers revised the proportion to an estimated 1.5–2.1%.[3] They also found that the Neanderthal component in non-African modern humans was more related to the Mezmaiskaya Neanderthal (Caucasus) than to the Altai Neanderthal (Siberia) or the Vindija Neanderthals (Croatia).[3] In the modern human population, at least those of East Asians and Europeans, the total introgressed Neanderthal DNA found spans about 20% of the Neanderthal genome.[4]
Although less parsimonious than recent gene flow, the observation may have been due to ancient population sub-structure in Africa, causing incomplete genetic homogenization within modern humans when Neanderthals diverged while early ancestors of Eurasians were still more closely related to Neanderthals than those of Africans to Neanderthals.[2] On the basis of allele frequency spectrum, it was shown that the recent admixture model had the best fit to the results while the ancient population sub-structure model had no fit–demonstrating that the best model was a recent admixture event that was preceded by a bottleneck event among modern humans—thus confirming recent admixture as the most parsimonious and plausible explanation for the observed excess of genetic similarities between modern non-African humans and Neanderthals.[5] A recent admixture event is likewise confirmed by data on the basis of linkage disequilibrium.[6]
Recent studies have shown a higher Neanderthal admixture in East Asians when compared to Europeans.[4][7][8][9] It indicated that most-likely at least two independent events of gene flow must have taken place into early modern humans and that the early ancestors of East Asians experienced more admixture than those of Europeans after the divergence of the two groups.[4][7][8] This is estimated to have been an additional 20.2% (95%CI of 13.4–27.1%) of Neanderthal admixture in a second gene flow to East Asians.[4] It is also possible, but less likely, that the difference was caused by dilution in Europeans by later migrations out of Africa.[7] It may also be due to a lower negative selection in East Asians compared to Europeans.[9] It has also been observed that there's a small but significant variation of Neanderthal admixture rates within European populations, but no significant variation within East Asian populations.[4]
A 2012 study found that North Africans have a Neanderthal admixture rate lying between that of Eurasians (highest) and Sub-Saharan Africans (lowest).[10] It has also shown a great variation within North Africans themselves, depending primarily on the amount of Eurasian versus sub-Saharan African ancestry.[10] However, there are indications that their Neanderthal admixture is not solely contributed by Eurasian introgression.[10]
Neanderthal contribution has been very scarcely but significantly found in the Maasai, an East African people.[8] After identifying African and non-African ancestry among the Maasai, it can be concluded that recent non-African modern human (post-Neanderthal) gene flow was the source of the contribution as about an estimated 30% of the Maasai genome can be traced to non-Africans from about 100 generations ago.[8]
Through the extent of linkage disequilibrium, it was estimated that the last Neanderthal gene flow into early ancestors of Europeans occurred 47,000–65,000 years BP (conservatively 37,000–86,000 years BP).[6] In conjunction with archaeological and fossil evidence, the gene flow is thought likely to have occurred somewhere in Western Eurasia, possibly the Middle East.[6]
No evidence of Neanderthal mitochondrial DNA has been found in modern humans.[11][12][13] This would suggest that successful admixture with Neanderthals happened paternally rather than maternally on the side of Neanderthals.[14][15] Possible hypotheses are that Neanderthal mtDNA had detrimental mutations that led to the extinction of carriers, that the hybrid offspring of Neanderthal mothers were raised in Neanderthal groups and became extinct with them, or that female Neanderthals and male Sapiens did not produce fertile offspring.[14]
Recent studies found the presence of large genomic regions with strongly reduced Neanderthal contribution in modern humans due to negative selection,[4][9] partly caused by hybrid male infertility.[9] These large regions of low Neanderthal contribution were most-pronounced on the X chromosome—with fivefold lower Neanderthal ancestry compared to autosomes—and contained relatively high numbers of genes specific to testes, meaning that modern humans have relatively few Neanderthal genes that are located on the X chromosome or expressed in the testes, consistent with the known fact that male infertility is affected by a disproportional large amount of genes on X chromosomes.[9] It has also been shown that Neanderthal ancestry has been selected against in conserved biological pathways, such as RNA processing.[9]
Genes affecting keratin were found to have been introgressed from Neanderthals into modern humans (shown in East Asians and Europeans), suggesting that these genes gave a morphological adaptation in skin and hair to modern humans to cope with non-African environments.[4][9] This is likewise for several genes involved in medical-relevant phenotypes, such as those affecting systemic lupus erythematosus, primary biliary cirrhosis, Crohn's disease, optic disk size, smoking behavior, interleukin 18 levels, and diabetes mellitus type 2.[9]
In a 2013 study, researchers found Neanderthal introgression of 18 genes—several of which are related to UV-light adaptation—within the chromosome 3p21.31 region (HYAL region) of East Asians.[16] The introgressive haplotypes were positively selected in only East Asian populations, rising steadily from 45,000 years BP until a sudden increase of growth rate around 5,000 to 3,500 years BP.[16] They occur at very high frequencies among East Asian populations in contrast to other Eurasian populations (e.g. European and South Asian populations).[16] The findings also suggests that this Neanderthal introgression occurred within the ancestral population shared by East Asians and Native Americans.[16]
Data from 2005 had previously shown that a group of alleles collectively know as haplogroup D of microcephalin, a critical regulatory gene for brain volume, originated from an archaic human population.[17] The results show that haplogroup D introgressed 37,000 years ago (based on the coalescence age of derived D alleles) into modern humans from an archaic human population that separated 1,1 million years ago (based on the separation time between D and non-D alleles), consistent with the period when Neanderthals and modern humans co-existed and diverged respectively.[17] The high frequency of the D haplogroup (70%) suggest that it was positively selected for in modern humans.[17] The distribution of the D allele of microcephalin is high outside Africa but low in sub-Saharan Africa, which further suggest that the admixture event happened in archaic Eurasian populations.[17] This distribution difference between Africa and Eurasia suggests that the D allele originated from Neanderthals.[18] However, a 2010 study found that a Neanderthal individual from the Mezzena Rockshelter (Monti Lessini, Italy) was homozygous for an ancestral allele of microcephalin, thus providing no support to the theory that Neanderthals contributed the D allele to modern humans but does not exclude a possibility of a Neanderthal origin of the D allele.[18] The 2010 Neanderthal genome study also could not confirm a Neanderthal origin of haplogroup D of the microcephalin gene.[2]
A 2011 study found that the immune system's HLA-A*02, A*26/*66, B*07, B*51, C*07:02, and C*16:02 was contributed from Neanderthals to modern humans.[19] After migrating out of Africa, modern humans encountered and interbred with archaic humans, which was advantageous for modern humans in rapidly restoring HLA diversity and acquiring new HLA variants that are better adapted to local pathogens.[19]
In 2015, Israel Hershkovitz of Tel Aviv University reported that a skull found in a cave in northern Israel, is "probably a woman, who lived and died in the region about 55,000 years ago, placing modern humans there and then for the first time ever" and pointing to a potential time and location when modern humans first interbred with Neanderthals.[20]
Morphology
According to a 1999 study, the early Upper Paleolithic burial remains of a modern human child from Abrigo do Lagar Velho (Portugal) featured traits indicating Neanderthal admixtures with modern humans dispersing into Iberia.[21] Considering the dating of the burial remains (24,500 years BP) and the persistence of Neanderthal traits long after the transitional period from a Neanderthal to a modern human population in Iberia (28,000–30,000 years BP), it was suggested that the child may have been a descendant of an already heavily-admixed population.[21]
In a 2006 study, researchers found that the early Upper Paleolithic modern human remains from Peştera Muierilor (Romania) of 35,000 years BP have the morphological pattern of European early modern humans, but possesses archaic and/or Neanderthal features, suggesting European early modern humans' admixture with rather than a full replacement of Neanderthals.[22] These features include a large interorbital breadth, a relatively flat superciliary arches, a prominent occipital bun, an asymmetrical and shallow mandibular notch shape, a high mandibular coronoid processus, the relative perpendicular mandibular condyle to notch crest position, and a narrow scapular glenoid fossa.[22]
In a 2003 study, researchers found that the early modern human Oase 1 mandible from Peștera cu Oase (Romania) of 34,000–36,000 14C years BP presented a mosaic of modern, archaic, and possible Neanderthal features.[23] It displayed a lingual bridging of the mandibular foramen, not present in earlier humans except Neanderthals of the late Middle and Late Pleistocene, thus suggesting affinity with Neanderthals.[23] Concluding from the Oase 1 mandible, there was apparently a significant craniofacial change of early modern humans of those from at least Europe, possibly due to some degree of admixture with Neanderthals.[23]
A 2007 study found that the earliest (before about 33 ka BP) European modern humans and the subsequent (Middle Upper Paleolithic) Gravettians, falling anatomically largely inline with the earliest (Middle Paleolithic) African modern humans, also had traits that are distinctively Neanderthal, suggesting that a solely Middle Paleolithic modern human ancestry was unlikely for European early modern humans.[24]
In March 2013, new data from the late-Neanderthal jaw from the Mezzena rockshelter (Monti Lessini, Italy) indicated possible interbreeding in late Italian Neanderthals.[25] The jaw (more specifically, a corpus mandibulae remnant) falls within the morphological range of modern humans, but also displayed strong similarities with some of the other Neanderthal specimens, indicating a change in late Neanderthal morphology due to possibly interbreeding with modern humans.[25]
History
The hypothesis, variously under the names of interbreeding, hybridization, admixture or hybrid-origin theory, has been discussed ever since the discovery of Neanderthal remains in the 19th century, though earlier writers believed that Neanderthals were a direct ancestor of modern humans. Thomas Huxley suggested that many Europeans bore traces of Neanderthal ancestry, but associated Neanderthal characteristics with primitivism, writing that since they "belong to a stage in the development of the human species, antecedent to the differentiation of any of the existing races, we may expect to find them in the lowest of these races, all over the world, and in the early stages of all races".[26]
Hans Peder Steensby in the 1907 article Racestudier i Danmark ("Race studies in Denmark") rejected that Neanderthals were ape-like or inferior, and, while emphasizing that all modern humans are of mixed origins, suggested interbreeding as the best available explanation of a significant number of observations which by then were available.[27]
In the early twentieth century, Carleton Coon argued that the Caucasoid race is of dual origin consisting of Upper Paleolithic (mixture of H. sapiens and H. neanderthalensis) types and Mediterranean (purely H. sapiens) types. He repeated his theory in his 1962 book The Origin of Races.[28]
Stan Gooch in Personality and Evolution (1973) and The Neanderthal Question (1977) develops a theory of Neanderthal/Cro-Magnon hybridization, based not on an examination of anatomy but of his understanding of modern human psychology and society, which he claimed owes a significant debt to Neanderthal culture. Gooch's theories were dismissed by academia. Gooch refined his theory in Cities of Dreams (1989) and The Neanderthal Legacy (2008).
Denisovans
A 2010 study has shown that Melanesians (e.g. Papua New Guinean and Bougainville Islander) share relative more alleles with Denisovans when compared to other of the studied Eurasians and Africans.[29] It estimated that 4% to 6% of the genome in Melanesians derives from Denisovans, while no other Eurasians or Africans displayed contributions of the Denisovan genes.[29] It has been observed that Denisovans contributed genes to Melanesians but not to East Asians, indicating that there was interaction between the early ancestors of Melanesians with Denisovans but that this interaction did not take place in the regions near southern Siberia, where as-of-yet the only Denisovan remains have been found.[29] In addition, a 2011 study has also shown a relative increased allele sharing between Denisovans and Aboriginal Australians, compared to other Eurasians and African populations, consistent with relative high admixture between early ancestors of Melanesians and Denisovans.[30]
In 2011, a study produced evidence that the highest presence of Denisovan admixture is in Oceanian populations, followed by many Southeast Asian populations, and none in East Asian populations.[31] There is significant Denisovan genetic material in eastern Southeast Asian and Oceanian populations (e.g. Aboriginal Australians, Near Oceanians, Polynesians, Fijians, eastern Indonesians, Philippine Mamanwa and Manobo), but not in certain western and continental Southeast Asian populations (e.g. western Indonesians, Malaysian Jehai, Andaman Onge, and mainland Asians), indicating that the Denisovan admixture event happened in Southeast Asia itself rather than mainland Eurasia.[31] The observation of high Denisovan admixture in Oceania and the lack thereof in mainland Asia suggests that early modern humans and Denisovans had interbred east of the Wallace Line that divides Southeast Asia.[32]
However, in contrast to the previous results, more-recent research found indications that mainland Asian and Native American populations had 0.2% Denisovan contribution, albeit twenty-five-fold lower than Oceanian populations.[3] The manner of gene flow to these populations is currently unknown.[3] After Oceanians, it has been observed that particularly Southeast Asians in general have affinity to Denisovans.[33]
Findings indicate that the Denisovan gene flow event happened to the common ancestors of Aboriginal Philippines, Aboriginal Australians, and New Guineans.[31][34] New Guineans and Australians have similar rates of Denisovan admixture, indicating that interbreeding with their common ancestors happened prior to their entry into Sahul (Pleistocene New Guinea and Australia), at least 44,000 years ago.[31] It has also been observed that the fraction of Near Oceanian ancestry in Southeast Asians is proportional to the Denisovan admixture, except in the Philippines where there is a higher proportional Denisovan admixture to Near Oceanian ancestry.[31] Reich et al. (2010) suggested a possible model of an early eastward migration wave of modern humans, some who were Philippine/New Guinean/Australian common ancestors that interbred with Denisovans, respectively followed by (1) divergence of the Philippine early ancestors, (2) interbreeding between the New Guinean and Australian early ancestors with a part of the same early-migration population that did not experience Denisovan gene flow, and (3) interbreeding between the Philippine early ancestors with a part of the population from a much-later eastward migration wave (the other part of which would become East Asians).[31]
It has been shown that Eurasians have some but significant lesser archaic-derived genetic material that overlaps with Denisovans, stemming from the fact that Denisovans are related to Neanderthals—who contributed to the Eurasian gene pool—rather than from interbreeding of Denisovans with the early ancestors of those Eurasians.[7][29]
The skeletal remains of an early modern human from the Tianyuan cave (near Zhoukoudian, China) of 40,000 years BP showed a Neanderthal contribution within the range of today's Eurasian modern humans, but it had no discernible Denisovan contribution.[35] It is ancestral to many Asian and Native American populations, but post-dated the divergence between Asians and Europeans.[35] The lack of a Denisovan component in the Tianyuan individual suggests that the genetic contribution had been always scarce in the mainland.[3]
A 2011 study, exploring the immune system's HLA alleles, suggested that HLA-B*73 introgressed from Denisovans into modern humans in western Asia due to the distribution pattern and divergence of HLA-B*73 from other HLA alleles.[19] In modern humans, HLA-B*73 is concentrated in western Asia, but it is rare or absent elsewhere.[19] Even though HLA-B*73 is not present in the sequenced Denisovan genome, the study noted that it was associated to the Denisovan-derived HLA-C*15:05 from the linkage disequilibrium, consistent with the estimated 98% of those modern humans who carried B*73 also carried C*15:05.[19]
The Denisovan's two HLA-A (A*02 and A*11) and two HLA-C (C*15 and C*12:02) allotypes correspond to common alleles in modern humans, whereas one of the Denisovan's HLA-B allotype corresponds to a rare recombinant allele and the other is absent in modern humans.[19] It is thought that these must have been contributed from Denisovans to modern humans, because it is unlikely to have been preserved independently in both for so long due to HLA alleles' high mutation rate.[19]
A 2014 study found that a EPAS1 gene variant was introduced from Denisovans to modern humans.[36] The ancestral variant upregulates hemoglobin levels to compensate for low oxygen levels—such as at high altitudes—but this also has the maladaption of increasing blood viscosity.[36] The Denisovan-derived variant on the other hand limits this increase of hemoglobin levels, thus resulting in a better altitude adaption.[36] The Denisovan-derived EPAS1 gene variant is common in Tibetans and was positively selected in their ancestors after they colonized the Tibetan plateau.[36]
Archaic African hominins
Rapid decay of fossils in African environments have made it currently unfeasible to compare modern human admixture with reference samples of archaic African hominins.[37]
In 2011, after finding three candidate regions with introgression by searching for unusual patterns of variations—indicating a different origin—in 61 non-coding regions from two hunter-gatherers (Biaka Pygmies and San, shown significant for admixture in the data) and one West African agricultural group (Mandinka, shown not significant for admixture in the data), researchers concluded that roughly 2% of the genetic material found in some Sub-Saharan African populations was inserted into the human genome approximately 35,000 years ago from archaic hominins that broke away from the modern human lineage around 700,000 years ago.[38] After a survey for the introgressive haplotypes across Sub-Saharan populations, it was suggested that the admixture event happened with archaic hominins that possibly once inhabited Central Africa.[38]
In 2012, researchers studied high-coverage whole-genome sequences of fifteen Sub-Saharan hunter-gatherer males from three groups—five Pygmies (three Baka, a Bedzan, and a Bakola) from Cameroon, five Hadza from Tanzania, and five Sandawe from Tanzania—finding signs that the ancestors of the hunter-gatherers interbred with one or more archaic human populations,[37] probably over 40,000 years ago.[39] They also found that the median time of the most recent common ancestor of the fifteen test subjects with the putative introgressive haplotypes was 1.2–1.3 mya.[37]
Alternative hypotheses
In a 2013 study, some researchers had suggested that the observed genetic affinities between archaic and modern human populations are mostly explained by common ancestral polymorphisms—and not admixture—followed by genetic drift, explaining that the differences in the observed genetic affinities among modern human populations are most-likely a result from different retention rates of these polymorphisms among the modern human populations.[40] However, they also stated that the study didn't rule out archaic introgression to modern humans.[40]
See also
References
- ↑ Stringer, C. (2012). "What makes a modern human". Nature 485 (7396): 33–35. doi:10.1038/485033a. PMID 22552077.
- ↑ 2.0 2.1 2.2 2.3 2.4 Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M. et al. (2010). "A Draft Sequence of the Neandertal Genome". Science 328 (5979): 710–722. doi:10.1126/science.1188021. PMID 20448178.
- ↑ 3.0 3.1 3.2 3.3 3.4 Prüfer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S. et al. (2014) [Online 2013]. "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature 505 (7481): 43–49. doi:10.1038/nature12886.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Vernot, B.; Akey, J. M. (2014). "Resurrecting Surviving Neandertal Lineages from Modern Human Genomes". Science 343 (6174): 1017–1021. doi:10.1126/science.1245938.
- ↑ Yang, M.A.; Malaspinas, A.S.; Durand, E.Y.; Slatkin, M. (2012). "Ancient Structure in Africa Unlikely to Explain Neanderthal and Non-African Genetic Similarity". Molecular Biology and Evolution 29 (10): 2987–2995. doi:10.1093/molbev/mss117.
- ↑ 6.0 6.1 6.2 Sankararaman, S.; Patterson, N.; Li, H.; Pääbo, S.; Reich, D; Akey, J.M. (2012). "The Date of Interbreeding between Neandertals and Modern Humans". PLoS Genetics 8 (10): e1002947. doi:10.1371/journal.pgen.1002947. PMC 3464203. PMID 23055938.
- ↑ 7.0 7.1 7.2 7.3 Meyer, M.; Kircher, M.; Gansauge, M.T.; Li, H.; Racimo, F.; Mallick, S. et al. (2012). "A High-Coverage Genome Sequence from an Archaic Denisovan Individual". Science 338 (6104): 222–226. doi:10.1126/science.1224344. PMC 3617501. PMID 22936568.
- ↑ 8.0 8.1 8.2 8.3 Wall, J.D.; Yang, M.A.; Jay, F.; Kim, S.K.; Durand, E.Y.; Stevison, L.S. et al. (2013). "Higher Levels of Neanderthal Ancestry in East Asians than in Europeans". Genetics 194 (1): 199–209. doi:10.1534/genetics.112.148213.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 Sankararaman, S.; Mallick, S.; Dannemann, M. Prüfer, K.; Kelso, J.; Pääbo, S. et al. (2014). "The genomic landscape of Neanderthal ancestry in present-day humans". Nature 507 (7492): 354–357. doi:10.1038/nature12961.
- ↑ 10.0 10.1 10.2 Sánchez-Quinto, F.; Botigué, L.R.; Civit, S.; Arenas, C.; Ávila-Arcos, M.C.; Bustamante, C.D. et al. (2012). "North African Populations Carry the Signature of Admixture with Neandertals". PLoS ONE 7 (10): e47765. doi:10.1371/journal.pone.0047765. PMC 3474783. PMID 23082212.
- ↑ Krings, M.; Stone, A.; Schmitz, R.W.; Krainitzki, H.; Stoneking, M.; Pääbo, Svante (1997). "Neandertal DNA Sequences and the Origin of Modern Humans". Cell 90 (1): 19–30. doi:10.1016/S0092-8674(00)80310-4. PMID 9230299.
- ↑ Serre, D.; Langaney, A.; Chech, M.; Teschler-Nicola, M.; Paunovic, M.; Mennecier, P. et al. (2004). "No Evidence of Neandertal mtDNA Contribution to Early Modern Humans". PLoS Biology 2 (3): 313–317. doi:10.1371/journal.pbio.0020057. PMC 368159. PMID 15024415.
- ↑ Wall, J.D.; Hammer, M.F. (2006). "Archaic admixture in the human genome". Current Opinion in Genetics & Development 16 (6): 606–610. doi:10.1016/j.gde.2006.09.006. PMID 17027252.
- ↑ 14.0 14.1 Mason, P.H.; Short, R.V. (2011). "Neanderthal-human Hybrids". Hypothesis 9 (1): e1. doi:10.5779/hypothesis.v9i1.215.
- ↑ Wang, C.C.; Farina, S.E.; Li, H. (2013) [Online 2012]. "Neanderthal DNA and modern human origins". Quaternary International 295: 126–129. doi:10.1016/j.quaint.2012.02.027.
- ↑ 16.0 16.1 16.2 16.3 Ding, Q.; Hu, Y.; Xu, S.; Wang, J.; Jin, L. (2014) [Online 2013]. "Neanderthal Introgression at Chromosome 3p21.31 was Under Positive Natural Selection in East Asians". Molecular Biology and Evolution 31 (3): 683–695. doi:10.1093/molbev/mst260.
- ↑ 17.0 17.1 17.2 17.3 Evans, P.D.; Mekel-Bobrov, N.; Vallender, E.J.; Hudson, R.R.; Lahn, B.T. (2006). "Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage". Proceedings of the National Academy of Sciences 103 (48): 18178–18183. doi:10.1073/pnas.0606966103. PMC 1635020. PMID 17090677.
- ↑ 18.0 18.1 Lari, M.; Rizzi, E.; Milani, L.; Corti, G.; Balsamo, C.; Vai, S. et al. (2010). "The Microcephalin Ancestral Allele in a Neanderthal Individual". PLoS ONE 5 (5): e10648. doi:10.1371/journal.pone.0010648.
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 19.6 Abi-Rached, L.; Jobin, M. J.; Kulkarni, S.; McWhinnie, A.; Dalva, K.; Gragert, L. et al. (2011). "The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans". Science 334 (6052): 89–94. doi:10.1126/science.1209202. PMC 3677943. PMID 21868630.
- ↑ Skull discovery suggests location where humans first had sex with Neanderthals
- ↑ 21.0 21.1 Duarte, C.; Maurício, J.; Pettitt, P.B.; Souto, P.; Trinkaus, E.; Plicht, H. van der; Zilhão, J. (1999). "The early Upper Paleolithic human skeleton from the Abrigo do Lagar Velho (Portugal) and modern-human emergence in Iberia". Proceedings of the National Academy of Sciences 96 (13): 7604–9. doi:10.1073/pnas.96.13.7604. PMC 22133. PMID 10377462.
- ↑ 22.0 22.1 Soficaru, A.; Dobos, A.; Trinkaus, E. (2006). "Early modern humans from the Peştera Muierii, Baia de Fier, Romania". Proceedings of the National Academy of Sciences 103 (46): 17196–201. doi:10.1073/pnas.0608443103. PMC 1859909. PMID 17085588.
- ↑ 23.0 23.1 23.2 Trinkaus E.; Moldovan O.; Milota S.; Bîlgăr A.; Sarcina L.; Athreya S. et al. (2003). "An early modern human from the Peştera cu Oase, Romania". Proceedings of the National Academy of Sciences 100 (20): 11231–6. doi:10.1073/pnas.2035108100. PMC 208740. PMID 14504393.
- ↑ Trinkaus, E. (2007). "European early modern humans and the fate of the Neandertals". Proceedings of the National Academy of Sciences 104 (18): 7367–72. doi:10.1073/pnas.0702214104. PMC 1863481. PMID 17452632.
- ↑ 25.0 25.1 Condemi, S.; Mounier, A.; Giunti, P.; Lari, M.; Caramelli, D.; Longo, L.; Frayer, D. (2013). "Possible Interbreeding in Late Italian Neanderthals? New Data from the Mezzena Jaw (Monti Lessini, Verona, Italy)". PLoS ONE 8 (3): e59781. doi:10.1371/journal.pone.0059781. PMC 3609795. PMID 23544098.
- ↑ Huxley, T. (1890). "The Aryan Question and Pre-Historic Man". Collected Essays: Volume VII, Man's Place in Nature.
- ↑ Steensby, H.P. (1907). "Racestudier i Danmark". Geografisk Tidsskrift 9: 135–145.
- ↑ Coon, C.S. (1962). The Origin of Races. p. 529.
- ↑ 29.0 29.1 29.2 29.3 Reich, D.; Green, R.E.; Kircher, M.; Krause, J.; Patterson, N.; Durand, E.Y. et al. (2010). "Genetic history of an archaic hominin group from Denisova Cave in Siberia". Nature 468 (7327): 1053–1060. doi:10.1038/nature09710. PMID 21179161.
- ↑ Rasmussen, M.; Guo, X.; Wang, Y.; Lohmueller, K.E.; Rasmussen, S.; Albrechtsen, A. et al. (2011). "An Aboriginal Australian Genome Reveals Separate Human Dispersals into Asia". Science 334 (6052): 94–98. doi:10.1126/science.1211177.
- ↑ 31.0 31.1 31.2 31.3 31.4 31.5 Reich, D.; Patterson, N.; Kircher, M.; Delfin, F.; Nandineni, M.R.; Pugach, I. et al. (2011). "Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania". The American Journal of Human Genetics 89 (4): 516–528. doi:10.1016/j.ajhg.2011.09.005. PMC 3188841. PMID 21944045.
- ↑ Cooper, A.; Stringer, C.B. (2013). "Did the Denisovans Cross Wallace's Line?". Science 342 (6156): 321–323. doi:10.1126/science.1244869.
- ↑ Skoglund, P.; Jakobsson, M. (2011). "Archaic human ancestry in East Asia". Proceedings of the National Academy of Sciences 108 (45): 18301–18306. doi:10.1073/pnas.1108181108.
- ↑ Flatow, I.; Reich, D. (31 August 2012). "Meet Your Ancient Relatives: The Denisovans". NPR.
- ↑ 35.0 35.1 Fu, Q.; Meyer, M.; Gao, X.; Stenzel, U.; Burbano, H.A.; Kelso, J.; Paabo, S. (2013). "DNA analysis of an early modern human from Tianyuan Cave, China". Proceedings of the National Academy of Sciences 110 (6): 2223–2227. doi:10.1073/pnas.1221359110.
- ↑ 36.0 36.1 36.2 36.3 Huerta-Sánchez, E.; Jin, X.; Asan; Bianba, Z.; Peter, B.M.; Vinckenbosch, N. et al. (2014). "Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA". Nature 512: 194–7. doi:10.1038/nature13408. PMID 25043035.
- ↑ 37.0 37.1 37.2 Lachance, J.; Vernot, B.; Elbers, C.C.; Ferwerda, B.; Froment, A.; Bodo, J.M. et al. (2012). "Evolutionary History and Adaptation from High-Coverage Whole-Genome Sequences of Diverse African Hunter-Gatherers". Cell 150 (3): 457–469. doi:10.1016/j.cell.2012.07.009.
- ↑ 38.0 38.1 Hammer, M.F.; Woerner, A.E.; Mendez, F.L.; Watkins, J.C.; Wall, J.D. (2011). "Genetic evidence for archaic admixture in Africa". Proceedings of the National Academy of Sciences 108 (37): 15123–15128. doi:10.1073/pnas.1109300108. PMC 3174671. PMID 21896735.
- ↑ Callaway, E. (26 July 2012). "Hunter-gatherer genomes a trove of genetic diversity". Nature. doi:10.1038/nature.2012.11076.
- ↑ 40.0 40.1 Lowery, R.K.; Uribe, G.; Jimenez, E.B.; Weiss, M.A.; Herrera, K.J.; Regueiro, M.; Herrera, R.J. (2013). "Neanderthal and Denisova genetic affinities with contemporary humans: Introgression versus common ancestral polymorphisms". Gene 530 (1): 83–94. doi:10.1016/j.gene.2013.06.005. PMID 23872234.