Aldose reductase
| Aldose reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 1.1.1.21 | ||||||||
| CAS number | 9028-31-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
In enzymology, aldose reductase (or aldehyde reductase) (EC 1.1.1.21) is a cytosolic NADPH-dependent oxidoreductase that catalyzes the reduction of a variety of aldehydes and carbonyls, including monosaccharides. It is primarily known for catalyzing the reduction of glucose to sorbitol, the first step in polyol pathway of glucose metabolism.[1]
Reactions
Aldose reductase catalyzes the NADPH-dependent conversion of glucose to sorbitol, the first step in polyol pathway of glucose metabolism. The second and last step in the pathway is catalyzed by sorbitol dehydrogenase, which catalyzes the NAD-linked oxidation of sorbitol to fructose. Thus, the polyol pathway results in conversion of glucose to fructose with stoichiometric utilization of NADPH and production of NADH.[1]
Galactose is also a substrate for the polyol pathway, but the corresponding keto sugar is not produced because sorbitol dehydrogenase is incapable of oxidizing galactitol.[2][2] Nevertheless, aldose reductase can catalyze the reduction of galactose to galactitol
- galactose + NADPH + H+
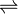 galactitol + NADP+
galactitol + NADP+
Function
The aldose reductase reaction, in particular the sorbitol produced, is important for the function of various organs in the body. For example, it is generally used as the first step in a synthesis of fructose from glucose; the second step is the oxidation of sorbitol to fructose catalyzed by sorbitol dehydrogenase. The main pathway from glucose to fructose (glycolysis) involves phosphorylation of glucose by hexokinase to form glucose 6-phosphate, followed by isomerization to fructose 6-phosphate and hydrolysis of the phosphate, but the sorbitol pathway is useful because it does not require the input of energy in the form of ATP:
- Seminal vesicles: Fructose produced from sorbitol is used by the sperm cells.
- Liver: Fructose produced from sorbitol can be used as an energy source for glycolysis and glyconeogenesis.
Aldose reductase is also present in the lens, retina, Schwann cells of peripheral nerves, placenta and red blood cells.
Enzyme Structure
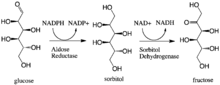
Aldose reductase may be considered a prototypical enzyme of the aldo-keto reductase enzyme superfamily. The enzyme comprises 315 amino acid residues and folds into a β/α-barrel structural motif composed of eight parallel β strands.[3] Adjacent strands are connected by eight peripheral α-helical segments running anti-parallel to the β sheet.[4] The catalytic active site situated in the barrel core.[4][5] The NADPH cofactor is situated at the top of the β/α barrel, with the nicotinamide ring projects down in the center of the barrel and pyrophosphate straddling the barrel lip.[1]
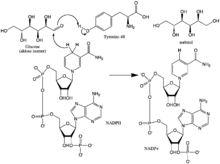
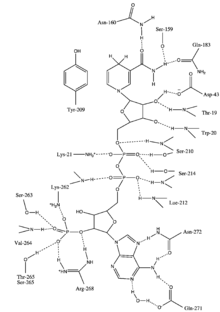


Enzyme Mechanism
The reaction mechanism of aldose reductase in the direction of aldehyde reduction follows a sequential ordered path where NADPH binds, followed by the substrate. Binding of NADPH induces a conformational change (Enzyme•NADPH –> Enzyme*•NADPH) that involves hinge-like movement of a surface loop (residues 213-217) so as to cover a portion of the NADPH in a manner similar to that of a safety belt. The alcohol product is formed via a transfer of the pro-R hydride of NADPH to the re face of the substrate's carbonyl carbon. Following release of the alcohol product, another conformational change occurs (E*•NADP+ –> E•NADP+) in order to release NADP+.[7] Kinetic studies have shown that reorientation of this loop to permit release of NADP+ appears to represent the rate-limiting step in the direction of aldehyde reduction.[8][9][10] As the rate of coenzyme release limits the catalytic rate, it can be seen that perturbation of interactions that stabilize coenzyme binding can have dramatic effects on the maximum velocity (Vmax).[10]
The hydride that is transferred from NADP+ to glucose comes from C-4 of the nicotinamide ring at the base of the hydrophobic cavity. Thus, the position of this carbon defines the enzyme's active site. There exist three residues in the enzyme within a suitable distance of the C-4 that could be potential proton donors: Tyr-48, His-110 and Cys-298. Evolutionary, thermodynamic and molecular modeling evidence predicted Tyr-48 as the proton donor. This prediction was confirmed the results of mutagenesis studies.[4][11][12] Thus, a [hydrogen-bonding] interaction between the phenolic hydroxyl group of Tyr-48 and the ammonium side chain of Lys-77 is thought to help to facilitate hydride transfer.[4]
Role in diabetes
Diabetes mellitus is recognized as a leading cause of new cases of blindness, and is associated with increased risk for painful neuropathy, heart disease and kidney failure. Many theories have been advanced to explain mechanisms leading to diabetic complications, including stimulation of glucose metabolism by the polyol pathway. Additionally, the enzyme is located in the eye (cornea, retina, lens), kidney, and the myelin sheath–tissues that are often involved in diabetic complications.[13] Under normal glycemic conditions, only a small fraction of glucose is metabolized through the polyol pathway, as the majority is phosphorylated by hexokinase, and the resulting product, glucose-6-phosphate, is utilized as a substrate for glycolysis or pentose phosphate metabolism.[14][15] However, in response to the chronic hyperglycemia found in diabetics, glucose flux through the polyol pathway is significantly increased. Up to 33% of total glucose utilization in some tissues can be through the polyol pathway.[16] Glucose concentrations are often elevated in diabetics and aldose reductase has long been believed to be responsible for diabetic complications involving a number of organs. Many aldose reductase inhibitors have been developed as drug candidates but virtually all have failed although some such as epalrestat are commercially available in several countries. Additional reductase inhibitors such as ranirestat, ponalrestat, rinalrestat, risarestat, sorbinil, and berberine[17] are currently in clinical trials.[18]
See also
References
- ↑ 1.0 1.1 1.2 Petrash JM (April 2004). "All in the family: aldose reductase and closely related aldo-keto reductases". Cell. Mol. Life Sci. 61 (7–8): 737–49. doi:10.1007/s00018-003-3402-3. PMID 15094999.
- ↑ 2.0 2.1 Jedziniak JA, Yates EM, Kinoshita JH (June 1973). "Lens polyol dehydrogenase". Exp. Eye Res. 16 (2): 95–104. doi:10.1016/0014-4835(73)90304-7. PMID 4352688. Retrieved 2010-05-18.
- ↑ Barski OA, Gabbay KH, Bohren KM (September 1999). "Characterization of the human aldehyde reductase gene and promoter". Genomics 60 (2): 188–98. doi:10.1006/geno.1999.5915. PMID 10486210. Retrieved 2010-05-18.
- ↑ 4.0 4.1 4.2 4.3 Wilson DK, Bohren KM, Gabbay KH, Quiocho FA (July 1992). "An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications". Science 257 (5066): 81–4. doi:10.1126/science.1621098. PMID 1621098. Retrieved 2010-05-18.
- ↑ Rondeau JM, Tête-Favier F, Podjarny A et al. (January 1992). "Novel NADPH-binding domain revealed by the crystal structure of aldose reductase". Nature 355 (6359): 469–72. doi:10.1038/355469a0. PMID 1734286.
- ↑ 6.0 6.1 Figure 11-4 in: Rod Flower; Humphrey P. Rang; Maureen M. Dale; Ritter, James M. (2007). Rang & Dale's pharmacology. Edinburgh: Churchill Livingstone. ISBN 0-443-06911-5.
- ↑ Nakano T, Petrash JM (August 1996). "Kinetic and spectroscopic evidence for active site inhibition of human aldose reductase". Biochemistry 35 (34): 11196–202. doi:10.1021/bi9608121. PMID 8780524.
- ↑ Grimshaw CE, Shahbaz M, Putney CG (October 1990). "Mechanistic basis for nonlinear kinetics of aldehyde reduction catalyzed by aldose reductase". Biochemistry 29 (42): 9947–55. doi:10.1021/bi00494a027. PMID 2125486.
- ↑ Kubiseski TJ, Hyndman DJ, Morjana NA, Flynn TG (April 1992). "Studies on pig muscle aldose reductase. Kinetic mechanism and evidence for a slow conformational change upon coenzyme binding". J. Biol. Chem. 267 (10): 6510–7. PMID 1551865. Retrieved 2010-05-18.
- ↑ 10.0 10.1 Grimshaw CE, Bohren KM, Lai CJ, Gabbay KH (November 1995). "Human aldose reductase: rate constants for a mechanism including interconversion of ternary complexes by recombinant wild-type enzyme". Biochemistry 34 (44): 14356–65. doi:10.1021/bi00044a012. PMID 7578039.
- ↑ Tarle I, Borhani DW, Wilson DK, Quiocho FA, Petrash JM (December 1993). "Probing the active site of human aldose reductase. Site-directed mutagenesis of Asp-43, Tyr-48, Lys-77, and His-110". J. Biol. Chem. 268 (34): 25687–93. PMID 8245005. Retrieved 2010-05-18.
- ↑ Bohren KM, Grimshaw CE, Lai CJ et al. (March 1994). "Tyrosine-48 is the proton donor and histidine-110 directs substrate stereochemical selectivity in the reduction reaction of human aldose reductase: enzyme kinetics and crystal structure of the Y48H mutant enzyme". Biochemistry 33 (8): 2021–32. doi:10.1021/bi00174a007. PMID 8117659.
- ↑ Schrijvers BF, De Vriese AS, Flyvbjerg A (December 2004). "From hyperglycemia to diabetic kidney disease: the role of metabolic, hemodynamic, intracellular factors and growth factors/cytokines". Endocr. Rev. 25 (6): 971–1010. doi:10.1210/er.2003-0018. PMID 15583025. Retrieved 2010-05-18.
- ↑ Gabbay KH, Merola LO, Field RA (January 1966). "Sorbitol pathway: presence in nerve and cord with substrate accumulation in diabetes". Science 151 (3707): 209–10. doi:10.1126/science.151.3707.209. PMID 5907911. Retrieved 2010-05-18.
- ↑ Lindstad RI, McKinley-McKee JS (September 1993). "Methylglyoxal and the polyol pathway. Three-carbon compounds are substrates for sheep liver sorbitol dehydrogenase". FEBS Lett. 330 (1): 31–5. doi:10.1016/0014-5793(93)80913-F. PMID 8370454. Retrieved 2010-05-18.
- ↑ Cheng HM, González RG (April 1986). "The effect of high glucose and oxidative stress on lens metabolism, aldose reductase, and senile cataractogenesis". Metab. Clin. Exp. 35 (4 Suppl 1): 10–4. doi:10.1016/0026-0495(86)90180-0. PMID 3083198.
- ↑ Wu LY, Ma ZM, Fan XL, Zhao T, Liu ZH, Huang X, Li MM, Xiong L, Zhang K, Zhu LL, Fan M (November 2009). "The anti-necrosis role of hypoxic preconditioning after acute anoxia is mediated by aldose reductase and sorbitol pathway in PC12 cells". Cell Stress Chaperones 15 (4): 387–94. doi:10.1007/s12192-009-0153-6. PMC 3082650. PMID 19902381.
- ↑ Schemmel KE, Padiyara RS, D'Souza JJ (September 2009). "Aldose reductase inhibitors in the treatment of diabetic peripheral neuropathy: a review". J. Diabetes Complicat. 24 (5): 354–60. doi:10.1016/j.jdiacomp.2009.07.005. PMID 19748287.
Further reading
- Denise R., PhD. Ferrier (2005). Lippincott's Illustrated Reviews: Biochemistry (Lippincott's Illustrated Reviews). Hagerstown, Maryland: Lippincott Williams & Wilkins. p. 319. ISBN 0-7817-2265-9.
- Attwood MA, Doughty CC (December 1974). "Purification and properties of calf liver aldose reductase". Biochim. Biophys. Acta 370 (2): 358–68. doi:10.1016/0005-2744(74)90097-7. PMID 4216364.
- Boghosian RA, McGuinness ET (April 1979). "Affinity purification and properties of porcine brain aldose reductase". Biochim. Biophys. Acta 567 (2): 278–86. doi:10.1016/0005-2744(79)90113-x. PMID 36151.
| ||||||||||||||||||||||||||||||||||||||||