Affinity propagation
In statistics and data mining, affinity propagation (AP) is a clustering algorithm based on the concept of "message passing" between data points.[1] Unlike clustering algorithms such as k-means or k-medoids, AP does not require the number of clusters to be determined or estimated before running the algorithm. Like k-medoids, AP finds "exemplars", members of the input set that are representative of clusters.[1]
Algorithm
Let x1 through xn be a set of data points, with no assumptions made about their internal structure, and let s be a function that quantifies the similarity between any two points, such that s(xi, xj) > s(xi, xk) iff xi is more similar to xj than to xk.
The algorithm proceeds by alternating two message passing steps, to update two matrices:[1]
- The "responsibility" matrix R has values r(i, k) that quantify how well-suited xk is to serve as the exemplar for xi, relative to other candidate exemplars for xi.
- The "availability" matrix A contains values a(i, k) represents how "appropriate" it would be for xi to pick xk as its exemplar, taking into account other points' preference for xk as an exemplar.
Both matrices are initialized to all zeroes, and can be viewed as log-probability tables. The algorithm then performs the following updates iteratively:
- First, responsibility updates are sent around:
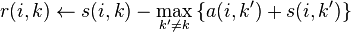
- Then, availability is updated per
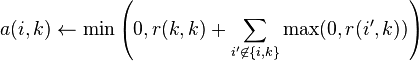 for
for 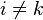 and
and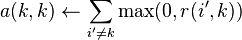 .
.
Applications
The inventors of affinity propagation showed it is better for certain computer vision and computational biology tasks, e.g. clustering of pictures of human faces and identifying regulated transcripts, than k-means,[1] even when k-means was allowed many random restarts and initialized using PCA.[2] A study comparing AP and Markov clustering on protein interaction graph partitioning found Markov clustering to work better for that problem.[3] A semi-supervised variant has been proposed for text mining applications.[4]
Software
- A Java implementation is included in the ELKI data mining framework.
- Java Apro library implements parallelized Affinity Propagation and Hierarchical AP.
- A Julia implementation of affinity propagation is contained in Julia Statistics's Clustering.jl package.[5]
- A Python version is part of the scikit-learn library.[6]
- An R implemenation is available in the "apcluster" package.[7]
References
- ↑ 1.0 1.1 1.2 1.3 Brendan J. Frey; Delbert Dueck (2007). "Clustering by passing messages between data points". Science 315 (5814): 972–976. doi:10.1126/science.1136800. PMID 17218491.
- ↑ Delbert Dueck; Brendan J. Frey (2007). Non-metric affinity propagation for unsupervised image categorization. Int'l Conf. on Computer Vision.
- ↑ James Vlasblom; Shoshana Wodak (2009). "Markov clustering versus affinity propagation for the partitioning of protein interaction graphs". BMC Bioinformatics 10 (1): 99. doi:10.1186/1471-2105-10-99.
- ↑ Renchu Guan; Xiaohu Shi; Maurizio Marchese; Chen Yang; Yanchun Liang (2011). "Text Clustering with Seeds Affinity Propagation". IEEE Transactions on Knowledge & Data Engineering 23 (4): 627–637.
- ↑ Clustering.jl www.github.com
- ↑ "Clustering — scikit-learn 0.14.1 documentation". Retrieved 15 July 2014.
- ↑ apcluster cran.r-project.org>