3DSlicer
 | |
| Original author(s) | The Slicer Community |
|---|---|
| Stable release | 4.4.0 / 25 December 2014 |
| Written in | C++, Python, Java, Qt |
| Operating system | Linux, Mac OS X, Windows |
| Size | 200MB |
| Available in | English |
| Type | Scientific visualization and image computing |
| License | BSD-style |
| Website |
www |
3D Slicer (Slicer) is a free and open source software package for image analysis[1] and scientific visualization. Slicer is used in a variety of medical applications, including autism, multiple sclerosis, systemic lupus erythematosus, prostate cancer, schizophrenia, orthopedic biomechanics, COPD, cardiovascular disease and neurosurgery.
About Slicer
3D Slicer is a free open source software (BSD-style license) that is a flexible, modular platform for image analysis and visualization. 3D Slicer can be extended to enable development of both interactive and batch processing tools for a variety of applications.
3D Slicer provides image registration, processing of DTI (diffusion tractography), an interface to external devices for image guidance support, and GPU-enabled volume rendering, among other capabilities. 3D Slicer has a modular organization that allows the addition of new functionality and provides a number of generic features not available in competing tools.
The interactive visualization capabilities of 3D Slicer include the ability to display arbitrarily oriented image slices, build surface models from image labels, and hardware accelerated volume rendering. 3D Slicer also supports a rich set of annotation features (fiducials and measurement widgets, customized colormaps).
Slicer's capabilities include:[2]
- Handling DICOM images and reading/writing a variety of other formats
- Interactive visualization of volumetric Voxel images, polygonal meshes, and volume renderings
- Manual editing
- Fusion and co-registering of data using rigid and non-rigid algorithms
- Automatic image segmentation
- Analysis and visualization of diffusion tensor imaging data
- Tracking of devices for image-guided procedures.
Slicer is compiled for use on multiple computing platforms, including Windows, Linux, and Mac OS X.
Slicer is distributed under a BSD style, free, open source license. The license has no restrictions on use of the software in academic or commercial projects. However, no claims are made on the software being useful for any particular task. It is entirely the responsibility of the user to ensure compliance with local rules and regulations. Slicer has not been formally approved for clinical use in the FDA in the US or by any other regulatory body elsewhere.
Image gallery
-

Hardware accelerated volume rendering with nVidia drivers, (on Windows and Linux only).
-

ProstateNav Module for MRI guided robot assisted biopsy of the prostate.
-

Left: 3D rendering. Right: Open MR system
-

Visualization of some atlas-based ROIs which correspond to major anatomical fiber tracts. The atlas was provided as part of a download of DTI studio.
-
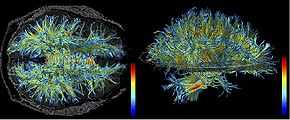
High resolution data acquired on 3-Tesla magnet and post-processed using automated tracking procedure.
-

High-dimensional white matter atlas generation and group analysis: result of automatic segmentation of novel subjects.
-
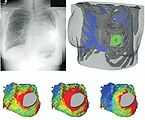
Patient-specific modeling in a patient with congenital heart disease.
-

Left: Three-dimensional model of levator ani subdivisions including the pubic bone and pelvic viscera. Right: The same model without the pubic bone.
-
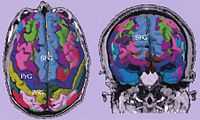
Cortical parcellations derived from SPGR images obtained from a tumor patient.
-

Intraoperative colocalization using iMRI images and 3-D Slicer software.
History
Slicer started as a masters thesis project between the Surgical Planning Laboratory at the Brigham and Women's Hospital and the MIT Artificial Intelligence Laboratory in 1998.[3] 3D Slicer version 2 has been downloaded several thousand times. In 2007 a completely revamped version 3 of Slicer was released. The next major refactoring of Slicer was initiated in 2009, which aims to transition the GUI of Slicer from using KWWidgets to Qt. Qt-enabled Slicer version 4 has been released in 2011.[4]
Slicer software has enabled a variety of research publications, all aimed at improving image analysis.[5]
This significant software project has been enabled by the participation of several large-scale NIH funded efforts, including the NA-MIC, NAC, BIRN, CIMIT, Harvard Catalyst and NCIGT communities. The funding support comes from several federal funding sources, including NCRR, NIBIB, NIH Roadmap, NCI, NSF and the DOD.
Users
Slicer's platform provides functionalities for segmentation, registration and three-dimensional visualization of multimodal image data, as well as advanced image analysis algorithms for diffusion tensor imaging, functional magnetic resonance imaging and image-guided radiation therapy. Standard image file formats are supported, and the application integrates interface capabilities to biomedical research software.
Slicer has been used in a variety of clinical research. In image-guided therapy research, Slicer is frequently used to construct and visualize collections of MRI data that are available pre- and intra-operatively to allow for the acquiring of spatial coordinates for instrument tracking.[6] In fact, Slicer has already played such a pivotal role in image-guided therapy, it can be considered as growing up alongside that field, with over 200 publications referencing Slicer since 1998.[7]
In addition to producing 3D models from conventional MRI images, Slicer has also been used to present information derived from fMRI (using MRI to assess blood flow in the brain related to neural or spinal cord activity),[8] DTI (using MRI to measure the restricted diffusion of water in imaged tissue),[9] and electrocardiography.[10] For example, Slicer's DTI package allows the conversion and analysis of DTI images. The results of such analysis can be integrated with the results from analysis of morphologic MRI, MR angiograms and fMRI. Other uses of Slicer include paleontology[11] and neurosurgery planning.[12]
Developers
The Slicer Developer Orientation offers resources for developers new to the platform. Slicer development is coordinated on the slicer-devel mailing list, and a summary of development statistics is available on Ohloh.
3D Slicer is built on VTK, a pipeline-based graphical library that is widely used in scientific visualization. In version 4, the core application is implemented in C++, and the API is available through a Python wrapper to facilitate rapid, iterative development and visualization in the included Python console. The user interface is implemented in Qt, and may be extended using either C++ or Python.
Slicer supports several types of modular development. Fully interactive, custom interfaces may be written in C++ or Python. Command-line programs in any language may be wrapped using a light-weight XML specification, from which a graphical interface is automatically generated.
For modules that are not distributed in the Slicer core application, a system is available to automatically build and distribute for selective download from within Slicer. This mechanism facilitates the incorporation of code with different license requirements from the permissive BSD-style license used for the Slicer core.
The Slicer build process utilizes CMake to automatically build prerequisite and optional libraries (excluding Qt). The core development cycle incorporates automatic testing, as well as incremental and nightly builds on all platforms, monitored using an online dashboard.
Criticism
Development still in progress, Slicer is sometimes accused by users of being poorly documented and a lacking in automation facilities (which is useful in batch processing). Other users report that Slicer has excellent documentation and training materials. Also Slicer's user interface and internal processing logic is fully scriptable. Although bugs can be reported to the mailing list and issue tracker, they are addressed based on developer availability. Updated versions are periodically released with updated features, while the development version with the latest source code is available daily.
External dependencies
See also
References
- ↑ Pieper S., Halle M., Kikinis R. 3D SLICER. Proceedings of the 1st IEEE International Symposium on Biomedical Imaging: From Nano to Macro 2004; 1:632–635.
- ↑ Pieper S., Lorensen B., Schroeder W., Kikinis R. The NA-MIC Kit: ITK, VTK, Pipelines, Grids and 3D Slicer as an Open Platform for the Medical Image Computing Community. Proceedings of the 3rd IEEE International Symposium on Biomedical Imaging: From Nano to Macro 2006; 1:698-701.
- ↑ Hirayasu, Y; Shenton, ME; Salisbury, DF; Dickey, CC; Fischer, IA; Mazzoni, P; Kisler, T; Arakaki, H; Kwon, JS; Anderson, JE; Yurgelun-Todd, D; Tohen, M; McCarley, RW (1998). "Lower left temporal lobe MRI volumes in patients with first-episode schizophrenia compared with psychotic patients with first-episode affective disorder and normal subjects". The American Journal of Psychiatry 155 (10): 1384–91. PMID 9766770.
- ↑ Fedorov; Beichel; Kalpathy-Cramer; Finet; Fillion-Robin; Pujol; Bauer; Jennings; Fennessy; Sonka; Buatti; Aylward; Miller; Pieper; Kikinis (2012). "3D Slicer as an image computing platform for the Quantitative Imaging Network". Magnetic Resonance Imaging 30 (9): 1323=41. doi:10.1016/j.mri.2012.05.001. PMID 22770690.
- ↑ Pieper S., Lorensen B., Schroeder W., Kikinis R. The NA-MIC Kit: ITK, VTK, Pipelines, Grids and 3D Slicer as an Open Platform for the Medical Image Computing Community. Proceedings of the 3rd IEEE International Symposium on Biomedical Imaging: From Nano to Macro 2006; 1:698–701.
- ↑ Hata, N; Piper, S; Jolesz, FA; Tempany, CM; Black, PM; Morikawa, S; Iseki, H; Hashizume, M; Kikinis, R (2007). "Application of open source image guided therapy software in MR-guided therapies". Medical image computing and computer-assisted intervention : MICCAI ... International Conference on Medical Image Computing and Computer-Assisted Intervention 10 (Pt 1): 491–8. doi:10.1007/978-3-540-75757-3_60. PMID 18051095.
- ↑ For a list of publications citing Slicer usage since 1998, visit: http://www.slicer.org/publications/pages/display/?collectionid=11
- ↑ Archip, N; Clatz, O; Whalen, S; Kacher, D; Fedorov, A; Kot, A; Chrisochoides, N; Jolesz, F; Golby, A; Black, PM; Warfield, SK (2007). "Non-rigid alignment of pre-operative MRI, fMRI, and DT-MRI with intra-operative MRI for enhanced visualization and navigation in image-guided neurosurgery". NeuroImage 35 (2): 609–24. doi:10.1016/j.neuroimage.2006.11.060. PMC 3358788. PMID 17289403.
- ↑ Ziyan, U; Tuch, D; Westin, CF (2006). "Segmentation of thalamic nuclei from DTI using spectral clustering". Medical image computing and computer-assisted intervention : MICCAI ... International Conference on Medical Image Computing and Computer-Assisted Intervention 9 (Pt 2): 807–14. doi:10.1007/11866763_99. PMID 17354847.
- ↑ Verhey, JF; Nathan, NS; Rienhoff, O; Kikinis, R; Rakebrandt, F; D'ambra, MN (2006). "Finite-element-method (FEM) model generation of time-resolved 3D echocardiographic geometry data for mitral-valve volumetry". Biomedical engineering online 5: 17. doi:10.1186/1475-925X-5-17. PMC 1421418. PMID 16512925.
- ↑ http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html
- ↑ http://picasaweb.google.com/107065747472066371420
External links
| Wikimedia Commons has media related to 3DSlicer. |
| ||||||||||
| ||||||||||||||||||||||||