Wobble base pair
In molecular biology, a wobble base pair is a non-Watson-Crick base pairing between two nucleotides in RNA molecules. The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C). Because hypoxanthine is the nucleobase of inosine, "I" is used for the former in order to maintain consistency of nucleic acid nomenclature, which otherwise follows the names of the nucleobases (e.g., "G" for both guanine and guanosine).[1] The thermodynamic stability of a wobble base pair is comparable to that of a Watson-Crick base pair. Wobble base pairs are fundamental in RNA secondary structure and are critical for the proper translation of the genetic code.
tRNA wobble
In the genetic code, there are 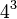 = 64 possible codons (tri-nucleotide sequences). For translation, each of these codons requires a tRNA molecule with a complementary anticodon. If each tRNA molecule paired with its complementary mRNA codon using canonical Watson-Crick base pairing, then 64 types (species) of tRNA molecule would be required. In the standard genetic code, three of these 64 codons are stop codons, which terminate translation by binding to release factors rather than tRNA molecules, so canonical pairing would require 61 species of tRNA. Since most organisms have fewer than 45 species of tRNA,[2] some tRNA species must pair with more than one codon. In 1966, Francis Crick proposed the Wobble hypothesis to account for this. He postulated that the 5' base on the anticodon, which binds to the 3' base on the mRNA, was not as spatially confined as the other two bases, and could, thus, have non-standard base pairing.[3] Movement ("wobble") of the base in the 5' anticodon position is necessary for this capability.[4]
= 64 possible codons (tri-nucleotide sequences). For translation, each of these codons requires a tRNA molecule with a complementary anticodon. If each tRNA molecule paired with its complementary mRNA codon using canonical Watson-Crick base pairing, then 64 types (species) of tRNA molecule would be required. In the standard genetic code, three of these 64 codons are stop codons, which terminate translation by binding to release factors rather than tRNA molecules, so canonical pairing would require 61 species of tRNA. Since most organisms have fewer than 45 species of tRNA,[2] some tRNA species must pair with more than one codon. In 1966, Francis Crick proposed the Wobble hypothesis to account for this. He postulated that the 5' base on the anticodon, which binds to the 3' base on the mRNA, was not as spatially confined as the other two bases, and could, thus, have non-standard base pairing.[3] Movement ("wobble") of the base in the 5' anticodon position is necessary for this capability.[4]
As an example, yeast tRNAPhe has the anticodon 5'-GmAA-3' and can recognize the codons 5'-UUC-3' and 5'-UUU-3'. It is, therefore, possible for non-Watson–Crick base pairing to occur at the third codon position, i.e., the 3' nucleotide of the mRNA codon and the 5' nucleotide of the tRNA anticodon.[5]
tRNA Base pairing schemes
The original wobble pairing rules, as proposed by Crick. Watson-Crick base pairs are shown in bold, wobble base pairs in italic:
| tRNA 5' anticodon base | mRNA 3' codon base |
|---|---|
| A | U |
| C | G |
| G | C or U |
| U | A or G |
| I | A or C or U |
Revised pairing rules[6]
| tRNA 5' anticodon base | mRNA 3' codon base |
|---|---|
| G | U,C |
| C | G |
| k2C | A |
| A | U,C,(A),G |
| unmodified U | U,(C),A,G |
| xm5s2U,xm5Um,Um,xm5U | A,(G) |
| xo5U | U,A,G |
| I | A or C or U |
See also
References
- ↑ Kuchin, Sergei (May 2011). "Covering All the Bases in Genetics: Simple Shorthands and Diagrams for Teaching Base Pairing to Biology Undergraduates". Journal of Microbiology & Biology Education (American Society for Microbiology) 12 (1). doi:10.1128/jmbe.v12i1.267. Retrieved October 16, 2013. "The correct name of the base in inosine (which is a nucleoside) is hypoxanthine, however, for consistency with the nucleic acid nomenclature, the shorthand [I] is more appropriate..."
- ↑ http://gtrnadb.ucsc.edu/
- ↑ Crick F (1966). "Codon–anticodon pairing: the wobble hypothesis". J Mol Biol 19 (2): 548–55. doi:10.1016/S0022-2836(66)80022-0. PMID 5969078.
- ↑ Mathews, Christopher; Van Holde, K.E.; Appling, Dean; Anthony-Cahill, Spencer (2013). Biochemistry (4th ed.). Toronto, Canada: Pearson Education. p. 1181. ISBN 978-0-13-800464-4.
- ↑ Varani G, McClain W (2000). "The G × U wobble base pair. A fundamental building-block of RNA structure crucial to RNA function in diverse biological systems". EMBO Rep 1 (1): 18–23. doi:10.1093/embo-reports/kvd001. PMC 1083677. PMID 11256617.
- ↑ Murphy FV 4th, Ramakrishnan V (Dec 2004). "Structure of a purine-purine wobble base pair in the decoding center of the ribosome". Nat Struct Mol Biol 11 (12): 1251–1252. doi:10.1038/nsmb866. PMID 15558050.
External links
- tRNA, the Adaptor Hypothesis and the Wobble Hypothesis
- Wobble base-pairing between codons and anticodons
