Turn (biochemistry)
A turn is an element of secondary structure in proteins where the polypeptide chain reverses its overall direction.
Definition
According to the most common definition,[1] a turn is a structural motif where the Cα atoms of two residues separated by few (usually 1 to 5) peptide bonds are in close approach (< 7 Å), while the corresponding residues do not form a regular secondary structure element such as an alpha helix or beta sheet. Contrary to helices, the backbone dihedral angles are not (roughly) constant for all the residues in the turn.
Although the close approach of the two terminal Cα atoms is usually correlated with the forming of one or two hydrogen bonds between the corresponding residues, such hydrogen bond is not strictly required in the definition of the turn. That said, in most cases the H-bonding and Cα-distance definitions are equivalent.
Types of turns
Tight turns
Turns are classified[2] according to the separation between the two end residues:
- In an α-turn the end residues are separated by four peptide bonds (
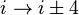 ).
). - In a β-turn (the most common form), by three bonds (
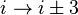 ).
). - In a γ-turn, by two bonds (
 ).
). - In a δ-turn, by one bond (
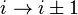 ).
). - In a π-turn, by five bonds (
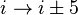 ).
).
| Type |  | 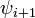 |  | 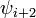 |
|---|---|---|---|---|
| I | -60 | -30 | -90 | 0 |
| II | -60 | 120 | 80 | 0 |
| VIII | -60 | -30 | -120 | 120 |
| I' | 60 | 30 | 90 | 0 |
| II' | 60 | -120 | -80 | 0 |
| VIa1 | -60 | 120 | -90 | 0* |
| VIa2 | -120 | 120 | -60 | 0* |
| VIb | -135 | 135 | -75 | 160* |
| IV |
turns excluded from all the above categories | |||
Within each type, turns may be further classified by their backbone dihedral angles (see Ramachandran plot). A turn can be converted into its inverse turn (also called its mirror-image turn) by changing the sign on all of its dihedral angles. (The inverse turn is not a true mirror image since the chirality of the Cα atoms is maintained.) Thus, the γ-turn has two forms, a classical form with (φ, ψ) dihedral angles of roughly (75°, -65°) and an inverse form with dihedral angles (-75°, 65°). At least eight forms of the β-turn have been identified, varying mainly in whether a cis isomer of a peptide bond is involved and on the dihedral angles of the central two residues. The classical and inverse β-turns are usually distinguished with a prime, e.g., type I and type I' β-turns.
Loops
An ω-loop is a catch-all term for a longer, extended or disordered loop without fixed internal hydrogen bonding.
Multiple turns
In many cases, one or more residues are involved in two partially overlapping turns. For example, in a sequence of 5 residues, both residues 1-4 and residues 2-5 form a turn; in such a case, one speaks of a  double turn. Multiple turns (up to 7-fold) occur in proteins, and they are found to be more common than single turns.[4]
double turn. Multiple turns (up to 7-fold) occur in proteins, and they are found to be more common than single turns.[4]
Hairpins
A hairpin is a special case of a turn, in which the direction of the protein backbone reverses and the flanking secondary structure elements interact. For example, a β-hairpin connects two hydrogen-bonded, antiparallel β-strands. (a rather confusing name, since a β-hairpin may contain many types of turns - α,β,γ, etc.)
β-hairpins may be classified according to the number of residues that make up the turn - that is, that are not part of the flanking β-strands.[5] If this number is X or Y (according to two different definitions of β sheets) the β hairpin is defined as X:Y
Role in protein folding
Two hypotheses have been proposed for the role of turns in protein folding. In one view, turns play a critical role in folding by bringing together and enabling or allowing interactions between regular secondary structure elements. This view is supported by mutagenesis studies indicating a critical role for particular residues in the turns of some proteins. Also, nonnative isomers of X-Pro peptide bonds in turns can completely block the conformational folding of some proteins. In the opposing view, turns play a passive role in folding. This view is supported by the poor amino-acid conservation observed in most turns. Also, non-native isomers of many X-Pro peptide bonds in turns have little or no effect on folding.
See also
- Secondary structure
External links
- NetTurnP - Prediction of Beta-turn regions in protein sequences
- BETATPRED - Prediction of Beta Turns in proteins using statistical algorithms
Notes
References
These references are ordered by date.
- Venkatachalam CM. (1968). "Stereochemical criteria for polypeptides and proteins. V. Conformation of a system of three linked peptide units". Biopolymers 6 (10): 1425–36. doi:10.1002/bip.1968.360061006. PMID 5685102.
- Némethy, George; Printz, Morton P. (1972). "The
 -Turn, a Possible Folded Conformation of the Polypeptide Chain. Comparison with the β-Turn". Macromolecules 5 (6): 755. doi:10.1021/ma60030a017.
-Turn, a Possible Folded Conformation of the Polypeptide Chain. Comparison with the β-Turn". Macromolecules 5 (6): 755. doi:10.1021/ma60030a017. - Lewis PN, Momany FA, Scheraga HA. (1973). "Chain reversals in proteins". Biochim Biophys Acta 303 (2): 211–29. PMID 4351002.
- Toniolo C.; Benedetti, Ettore (1980). "Intramolecularly hydrogen-bonded peptide conformations". CRC Crit Rev Biochem 9 (1): 1–44. doi:10.3109/10409238009105471. PMID 6254725.
- Richardson JS. (1981). "The anatomy and taxonomy of protein structure". Adv Protein Chem 34: 167–339. doi:10.1016/S0065-3233(08)60520-3. PMID 7020376.
- Rose GD, Gierasch LM, Smith JA. (1985). "Turns in peptides and proteins". Adv Protein Chem 37: 1–109. doi:10.1016/S0065-3233(08)60063-7. PMID 2865874.
- Milner-White EJ and Poet R. (1987). "Loops, bulges, turns and hairpins in proteins". Trends Biochem Sci 12: 189–192. doi:10.1016/0968-0004(87)90091-0.
- Wilmot CM, Thornton JM. (1988). "Analysis and prediction of the different types of beta-turn in proteins". J Mol Biol 203 (1): 221–32. doi:10.1016/0022-2836(88)90103-9. PMID 3184187.
- Sibanda, B.L.; Blundell, T.L.; Thornton, J.M. (1989). "Conformation of β-hairpins in protein structures:: A systematic classification with applications to modelling by homology, electron density fitting and protein engineering". Journal of Molecular Biology 206 (4): 759–777. doi:10.1016/0022-2836(89)90583-4. PMID 2500530. Retrieved 2011-02-15.
- Milner-White, E (1990). "Situations of gamma-turns in proteinsTheir relation to alpha-helices, beta-sheets and ligand binding sites". J. Mol. Biol. 216 (2): 385. doi:10.1016/S0022-2836(05)80329-8. PMID 2254936.
- Hutchinson, E.G.; Thornton, J.M. (1994). "A revised set of potentials for β-turn formation in proteins". Protein Science 3 (12): 2207–2216. doi:10.1002/pro.5560031206. PMC 2142776. PMID 7756980.
- Pavone V, Gaeta G, Lombardi A, Nastri F, Maglio O, Isernia C, Saviano M. (1996). "Discovering protein secondary structures: classification and description of isolated alpha-turns". Biopolymers 38 (6): 705–21. doi:10.1002/(SICI)1097-0282(199606)38:6<705::AID-BIP3>3.0.CO;2-V. PMID 8652792.
- Rajashankar KR, Ramakumar S. (1996). "Pi-turns in proteins and peptides: Classification, conformation, occurrence, hydration and sequence". Protein Sci 5 (5): 932–46. doi:10.1002/pro.5560050515. PMC 2143406. PMID 8732765.
| ||||||||||||||||||||||||||||||
 -turn types.
-turn types.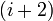 (*) must be a cis-
(*) must be a cis-