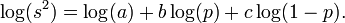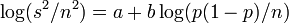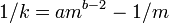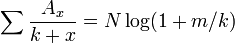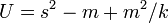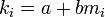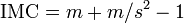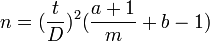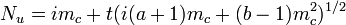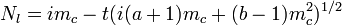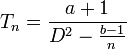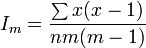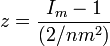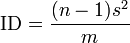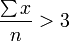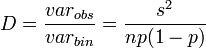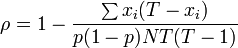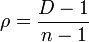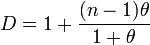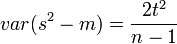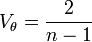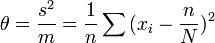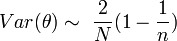Taylor's law
Taylor's law is an empirical law in ecology that relates the between-sample variance in density to the overall mean density of a sample of organisms in a study area.[1] Taylor described this relationship in 1961[2] and it has been found to be true for many species since.[3] It has also been found to be true in other areas including transmission of infectious diseases, human sexual behavior, childhood leukemia, cancer metastases, blood flow heterogeneity, genomic distributions of single nucleotide polymorphisms and gene structures.[4][5][6] This law is also known in the literature as the power law (in the biological literature) or the fluctuation scaling law (in the physics literature).
It is possible to derive this law if it is assumed that the organisms of interest form clusters that obey a Poisson distribution.[7] Alternative suggestions for its origin have also been proposed.[8]
History
The first to propose an empirical relationship of this type between the mean and variance was Smith in 1938 while studying crop yields.[9] Smith proposed the relationship
where Vx is the variance of yield for plots of x units, V1 is the variance of yield per unit area and x is the size of plots. The slope (b) is the index of heterogeneity. The value of b in this relationship lies between 0 and 1. Where the yield are highly correlated b tends to 0; when they are uncorrelated b tends to 1.
Fracker and Brischle in 1944[10] and Hayman and Lowe in 1961[11] independently described relationships between the mean and variance that are now known as Taylor's law.
The law itself is named after the ecologist L. R. Taylor (1924–2007). The name 'Taylor's law' was coined by Southwood in 1966.[12] Taylor's original name for this relationship was the law of the mean.
It appears that Taylor's law is an example of Stigler's law of eponymy.
Mathematical formulation
In symbols
where si2 is the variance of the density of the ith sample, mi is the mean density of the ith sample and a and b are constants.
In logarithmic form
Extensions and refinements
A refinement in the estimation of the slope b has been proposed by Rayner.[13]
where r is the Pearson moment correlation coefficient between log(s2) and log m, f is the ratio of sample variances in log(s2) and log m and φ is the ratio of the errors in log(s2) and log m.
Ordinary least squares regression assumes that φ = ∞. This tends to underestimate the value of b because the estimates of both log(s2) and log m are subject to error.
A extension of Taylor's law has been proposed by Ferris et al when multiple samples are taken[14]
where s2 and m are the variance and mean respectively, b, c and d are constants and n is the number of samples taken. To date this proposed extension has not been verified to be as applicable as the original version of Taylor's law.
Interpretation
Slope values (b) significantly > 1 indicate clumping of the organisms.
In Poisson distributed data b = 1.[15] If the population follows a lognormal or gamma distribution then b = 2.
Populations that are experiencing constant per capita environmental variability the regression of log( variance ) versus log( mean abundance ) should have a line with b = 2.
Most populations that have been studied have b < 2 (usually 1.5–1.6) but values of 2 have been reported.[5] Occasionally cases with b > 2 have been reported.[16] b values below 1 are uncommon but have also been reported ( b = 0.93 ).[17]
Notes
The origin of the slope (b) in this regression remains unclear. Two hypotheses have been proposed to explain it. One suggests that b arises from the species behavior and is a constant for that species. The alternative suggests that it is dependent on the sampled population. Despite the considerable number of studies carried out on this law (>1000) this question remains open.
It is known that both a and b are subject to change due to age-specific dispersal, mortality and sample unit size.[18]
This law may be a poor fit if the values are small. For this reason an extension to Taylor's law has been proposed by Hanski which improves the fit of Taylor's law at low densities.[19]
Extension to binary sampling
Binomial sampling is popular where there are large number of units (crops, trees) to be examined and where counts of individuals of interest (typically insects) may be difficult (frequently because the insects fly away before they can be accurately counted).
A form of Taylor's law applicable to binary sampling (presence/absence of at least one individual in a sample unit) has been proposed.[20] In a binomial distribution the theoretical variance is
where s2 is the variance, n is the sample size and p is the proportion of sample units with at least one individual. The proposed binary form of Taylor's law is
where varobs is the observed variance and varbin is that expected from the binomial distribution. When both a and b are equal to 1, then a random spatial pattern is suggested and is best described by the binomial distribution. When b = 1 and a > 1, there is overdispersion with no dependence on the mean incidence (p). When both a and b are > 1, the degree of aggregation varies with p.
Applications
Because of the ubiquitous occurrence of Taylor's law in biology it has found a variety of uses some of which are listed here.
Recommendations as to use
It has been recommended based on simulation studies[21] in applications testing the validity of Taylor's law to a data sample that:
(1) the total number of organisms studied be > 15
(2) the minimum number of groups of organisms studied be > 5
(3) the density of the organisms should vary by at least 2 orders of magnitude within the sample
Randomly distributed populations
It is common assumed (at least initially) that a population is randomly distributed in the environment. If a population is randomly distributed then the mean ( m ) and variance ( s2 ) of the population are equal and the proportion of samples that contain at least one individual ( p ) is
When a species with a clumped pattern is compared with one that is randomly distributed with equal overall densities, p will be less for the species having the clumped distribution pattern. Conversely when comparing a uniformly and a randomly distributed species but at equal overall densities, p will be greater for the randomly distributed population. This can be graphically tested by plotting p against m.
Wilson and Room developed a binomial model that incorporates Taylor's law.[22] The basic relationship is
where the log is taken to the base e.
Incorporating Taylor's law this relationship becomes
Dispersion parameter estimator
The common dispersion parameter (k) of the negative binomial distribution is
where m is the sample mean and s2 is the variance.[23] If 1 / k is > 0 the population is considered to be aggregated; 1 / k = 0 ( s2 = m ) the population is considered to be randomly (Poisson) distributed and if 1 / k is < 0 the population is considered to be uniformly distributed. No comment on the distribution can be made if k = 0.
Wilson and Room assuming that Taylor's law applied to the population gave an alternative estimator for k:[22]
where a and b are the constants from Taylor's law.
Jones[24] using the estimate for k above along with the relationship Wilson and Room developed for the probability of finding a sample having at least one individual[22]
derived an estimator for the probability of a sample containing x individuals per sampling unit. Jones's formula is
where P( x ) is the probability of finding x individuals per sampling unit, k is estimated from the Wilon and Room equation and m is the sample mean. The probability of finding zero individuals P( 0 ) is estimated with the negative binomial distribution
Jones also gives confidence intervals for these probabilities.
where CI is the confidence interval, t is the critical value taken from the t distribution and N is the total sample size.
Katz family of distributions
Katz proposed a family of distributions (the Katz family) with 2 parameters ( w1, w2 ).[25] This family of distributions includes the Bernoulli, Geometric, Pascal and Poisson distributions as special cases. The mean and variance of a Katz distribution are
where m is the mean and s2 is the variance of the sample. The parameters can be estimated by the method of moments from which we have
For a Poisson distribution w2 = 0 and w1 = λ the parameter of the Possion distribution. This family of distributions is also sometimes known as the Panjer family of distributions.
The Katz family is related to the Sundt-Jewel family of distributions:[26]
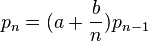
The only members of the Sundt-Jewel family are the Poisson, binomial, negative binomial (Pascal), extended truncated negative binomial and logarithmic series distributions.
If the population obeys a Katz distribution then the coefficients of Taylor's law are
Katz also introduced a statistical test[25]
where Jn is the test statistic, s2 is the variance of the sample, m is the mean of the sample and n is the sample size. Jn is asymptotically normally distributed with a zero mean and unit variance. If the sample is Poisson distributed Jn = 0; values of Jn < 0 and > 0 indicate under and over dispersion respectively. Overdispersion is often caused by latent heterogeneity - the presence of multiple sub populations within the population the sample is drawn from.
If the population obeys Taylor's law then
Time to extinction
If Taylor's law is assumed to apply it is possible to determine the mean time to local extinction. This model assumes a simple random walk in time and the absence of density dependent population regulation.[27]
Let 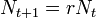 where Nt+1 and Nt are the population sizes at time t + 1 and t respectively and r is parameter equal to the annual increase (decrease in population). Then
where Nt+1 and Nt are the population sizes at time t + 1 and t respectively and r is parameter equal to the annual increase (decrease in population). Then
where var( r ) is the variance of r.
Let K be a measure of the species abundance (organisms per unit area). Then
where TE is the mean time to local extinction.
The probability of extinction by time t is
Minimum population size required to avoid extinction
If a population is lognormally distributed then the harmonic mean of the population size (H) is related to the arithmetic mean (m)[28]
Given that H must be > 0 for the population to persist then rearranging we have
is the minimum size of population for the species to persist.
The assumption of a lognormal distribution appears to apply to about half of a sample of 544 species.[29] suggesting that it is at least a plausible assumption.
Sampling size estimators
The degree of precision (D) is defined to be s / m where s is the standard deviation and m is the mean. The degree of precision is known as the coefficient of variation in other contexts. In ecology research it is recommended that D be in the range 10-25%.[30] The desired degree of precision is important in estimating the required sample size where an investigator wishes to test if Taylor's law applies to the data. The required sample size has been estimated for a number of simple distributions but where the population distribution is not known or cannot be assumed more complex formulae may needed to determine the required sample size.
Where the population is Poisson distributed the sample size (n) needed is
where t is critical level of the t distribution for the type 1 error with the degrees of freedom that the mean (m) was calculated with.
If the population is distributed as a negative binomial distribution then the required sample size is
where k is the parameter of the negative binomial distribution.
A more general sample size estimator has also been proposed[31]
where a and b are derived from Taylor's law.
An alternative has been proposed by Southwood[32]
where n is the required sample size, a and b are the Taylor's law coefficients and D is the desired degree of precision.
Karandinos proposed two similar estimators for n.[33] The first was modified by Ruesink to incorporate Taylor's law.[34]
where d is the ratio of half the desired confidence interval (CI) to the mean. In symbols
The second estimator is used in binomial (presence-absence) sampling. The desired sample size (n) is
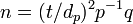
where the dp is ratio of half the desired confidence interval to the proportion of sample units with individuals, p is proportion of samples containing individuals and q = 1 - p. In symbols
Sequential sampling
Sequential analysis is a method of statistical analysis where the sample size is not fixed in advance. Instead samples are taken in accordance with a predefined stopping rule. Taylor's law has been used to derive a number of stopping rules.
A formula for fixed precision in serial sampling to test Taylor's law was derived by Green in 1970.[35]
where T is the cumulative sample total, D is the level of precision, n is the sample size and a and b are obtained from Taylor's law.
As an aid to pest control Wilson et al developed a test that incorporated a threshold level where action should be taken.[36] The required sample size is
where a and b are the Taylor coefficients, || is the absolute value, m is the sample mean, T is the threshold level and t is the critical level of the t distribution. The authors also provided a similar test for binomial (presence-absence) sampling
where p is the probability of finding a sample with pests present and q = 1 - p.
Green derived another sampling formula for sequential sampling based on Taylor's law[37]
where D is the degree of precision, a and b are the Taylor's law coefficients, n is the sample size and T is the total number of individuals sampled.
Related analyses
It is considered to be good practice to estimate at least one additional analysis of aggregation (other than Taylor's law) because the use of only a single index may be misleading.[38] Although a number of other methods for detecting relationships between the variance and mean in biological samples have been proposed, to date none have achieved the popularity of Taylor's law. The most popular analysis used in conjunction with Taylor's law is probably Iowa's Patchiness regression test but all the methods listed here have been used in the literature.
Barlett-Iawo model
Barlett in 1936[39] and later Iawo independently in 1968[40] both proposed an alternative relationship between the variance and the mean. In symbols
where s is the variance in the ith sample and mi is the mean of the ith sample
When the population follows a negative binomial distribution, a = 1 and b = k (the exponent of the negative binomial distribution).
This alternative formulation has not been found to be as good a fit as Taylor's law in most studies.
Nachman model
Nachman proposed a relationship between the mean density and the proportion of samples with zero counts:[41]
where p0 is the proportion of the sample with zero counts, m is the mean density, a is a scale parameter and b is a dispersion parameter. If a = b = 0 the distribution is random. This relationship is usually tested in its logarithmic form
Kono-Sugino equation
Binary sampling is not uncommonly used in ecology. In 1958 Kono and Sugino derived an equation that relates the proportion of samples without individuals to the mean density of the samples.[42]
where p0 is the proportion of the sample with no individuals, m is the mean sample density, a and b are constants. Like Taylor's law this equation has been found to fit a variety of populations including ones that obey Taylor's law. Unlike the negative binomial distribution this model is independent of the mean density.
- Note
The equation was derived while examining the relationship between the proportion ( P ) of a series of rice hills infested and the mean severity of infestation ( m ). The model studied was
where a and b are empirical constants. Based on this model the constants a and b were derived and a table prepared relating the values of P and m
- Uses
The predicted estimates of m from this equation are subject to bias[43] and it is recommended that the adjusted mean ( ma ) be used instead[44]
where var() is the variance of the sample unit means ( mi ) and m is the overall mean.
An alternative adjustment to the mean estimates is[44]
where MSE is the mean square error of the regression.
This model may also be used to estimate stop lines for enumerative (sequential) sampling. The variance of the estimated means is[45]
where
where MSE is the mean square error of the regression, α and β are the constant and slope of the regression respectively, sβ2 is the variance of the slope of the regression, N is the number of points in the regression, n is the number of sample units and p is the mean value of p0 in the regression. The parameters a and b are estimated from Taylor's law:
Hughes-Madden equation
Hughes and Madden have proposed testing a similar relationship also applicable to binary sampling (presence/absence in a sampled unit)[46][47]
where a, b and c are constants, s2 is the variance and p is the proportion of units with at least one individual. In logarithmic form this relationship is
This relationship has not yet been subjected to the extensive testing that Taylor's law has been subjected to. For this reason its general applicability presently remains uncertain.
A variant of this equation was proposed by Shiyomi et al[48] who suggested testing the regression
where s2 is the variance, a and b are the constants of the regression, n is the sample size and p is the probability of a sample containing at least one individual.
Negative binomial distribution model
A negative binomial model has also been proposed.[49] The dispersion parameter (k) using the method of moments is m2 / ( s2 - m ) and pi is the proportion of samples with counts > 0. The s2 used in the calculation of k are the values predicted by Taylor's law. pi is plotted against 1 - ( k ( k + m ) −1 )k and the fit of the data is visually inspected.
Perry and Taylor have proposed an alternative estimator of k based on Taylor's law.[50]
A better estimate of the dispersion parameter can be made with the method of maximum likelihood. For the negative binomial it can be estimated from the equation[23]
where Ax is the total number of samples with more than x individuals, N is the total number of individuals, x is the number of individuals in a sample, m is the mean number of individuals per sample and k is the exponent. The value of k has to estimated numerically.
Goodness of fit of this model can be tested in a number of ways including using the chi square test. As these may be biased by small samples an alternative is the U statistic - the difference between the variance expected under the negative binomial distribution and that of the sample. The expected variance of this distribution is m + m2 / k and
where s2 is the sample variance, m is the sample mean and k is the negative binomial parameter.
The variance of U is[23]
where p = m / k, q = 1 + p, R = p / q and N is the total number of individuals in the sample. The expected value of U is 0. For large sample sizes U is distributed normally.
Tests for a common dispersion parameter
The dispersion parameter (k)[23] is
where m is the sample mean and s2 is the variance. If k−1 is > 0 the population is considered to be aggregated; k−1 = 0 the population is considered to be random; and if k−1 is < 0 the population is considered to be uniformly distributed.
Southwood has recommended regressing k against the mean and a constant[32]
where ki and mi are the dispersion parameter and the mean of the ith sample respectively to test for the existence of a common dispersion parameter (kc). A slope (b) value significantly > 0 indicates the dependence of k on the mean density.
An alternative method was proposed by Elliot who suggested plotting ( s2 - m ) against ( m2 - s2 / n ).[51] kc is equal to 1/slope of this regression.
Lloyd's indexes
Lloyd's index of mean crowding (IMC) is the average number of other points contained in the sample unit that contains a randomly chosen point.[52]
where m is the sample mean and s2 is the variance.
Lloyd's index of patchiness (IP)[52] is
It is a measure of pattern intensity that is unaffected by thinning (random removal of points). This index was also proposed by Pielou in 1988 and is sometimes known by this name also.
If the population obeys Taylor's law then
Patchiness regression test
Iwao proposed a patchiness regression to test for clumping[40][53]
Let
yi here is Lloyd's index of mean crowding.[52] Perform an ordinary least squares regression of mi against y.
In this regression the value of the slope (b) is an indicator of clumping: the slope = 1 if the data is Poisson-distributed. The constant (a) is the number of individuals that share a unit of habitat at infinitesimal density and may be < 0, 0 or > 0. These values represent regularity, randomness and aggregation of populations in spatial patterns respectively. A value of a < 1 is taken to mean that the basic unit of the distribution is a single individual.
Where the statistic s2 / m is not constant it has been recommended to use instead to regress Lloyd's index against am + bm2 where a and b are constants.[54]
The sample size (n) for a given degree of precision (D) for this regression is given by[54]
where a is the constant in this regression, b is the slope, m is the mean and t is the critical value of the t distribution.
Iawo has proposed a sequential sampling test based on this regression.[55] The upper and lower limits of this test are based on critical densities mc where control of a pest requires action to be taken.
where Nu and Nl are the upper and lower bounds respectively, a is the constant from the regression, b is the slope and i is the number of samples.
Kuno has proposed an alternative sequential stopping test also based on this regression.[56]
where Tn is the total sample size, D is the degree of precision, n is the number of samples units, a is the constant and b is the slope from the regression respectively.
Kuno's test is subject to the condition that n ≥ (b - 1) / D2
Parrella and Jones have proposed an alternative but related stop line[57]
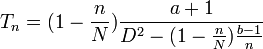
where a and b are the parameters from the regression, N is the maximum number of sampled units and n is the individual sample size.
Morisita’s index of dispersion
Morisita’s index of dispersion ( Im ) is the scaled probability that two points chosen at random from the whole population are in the same sample.[58] Higher values indicate a more clumped distribution.
An alternative formulation is
where n is the total sample size, m is the sample mean and x are the individual values with the sum taken over the whole sample. It is also equal to
where IMC is Lloyd's index of crowding.[52]
This index is relatively independent of the population density but is affected by the sample size.
Morisita showed that the statistic[58]
is distributed as a chi squared variable with n - 1 degrees of freedom.
A alternative significance test for this index has been developed for large samples.[59]
where m is the overall sample mean, n is the number of sample units and z is the normal distribution abscissa. Significance is tested by comparing the value of z against the values of the normal distribution.
A function for its calculation is available in the statistical R language. R function
Standardised Morisita’s index
Smith-Gill developed a statistic based on Morisita’s index which is independent of both sample size and population density and bounded by -1 and +1. This statistic is calculated as follows[60]
First determine Morisita's index ( Id ) in the usual fashion. Then let k be the number of units the population was sampled from. Calculate the two critical values
where χ2 is the chi square value for n - 1 degrees of freedom at the 97.5% and 2.5% levels of confidence.
The standardised index ( Ip ) is then calculated from one of the formulae below
When Id ≥ Mc > 1
When Mc > Id ≥ 1
When 1 > Id ≥ Mu
When 1 > Mu > Id
Ip ranges between +1 and -1 with 95% confidence intervals of ±0.5. Ip has the value of 0 if the pattern is random; if the pattern is uniform, Ip < 0 and if the pattern shows aggregation, Ip > 0.
Southwood's index of spatial aggregation
Southwood's index of spatial aggregation (k) is defined as
where m is the mean of the sample and m* is Lloyd's index of crowding.[32]
Fisher's index of dispersion
Fisher's index of dispersion[51][61] is
This index may be used to test for over dispersion of the population. It is recommended that in applications n > 5[62] and that the sample total divided by the number of samples is > 3. In symbols
where x is an individual sample value. The expectation of the index is equal to n and it is distributed as the chi-square distribution with n − 1 degrees of freedom when the population is Poisson distributed.[62] It is equal to the scale parameter when the population obeys the gamma distribution.
It can be applied both to the overall population and to the individual areas sampled individually. The use of this test on the individual sample areas should also include the use of a Bonferroni correction factor.
If the population obeys Taylor's law then
Index of cluster size
The index of cluster size (ICS) was created by David and Moore.[63] Under a random (Poisson) distribution ICS is expected to equal 0. Positive values indicate a clumped distribution; negative values indicate a uniform distribution.
where s2 is the variance and m is the mean.
If the population obeys Taylor's law
The ICS is also equal to Katz's test statistic divided by ( n / 2 )1/2 where n is the sample size. It is also related to Clapham's test statistic. It is also sometimes referred to as the clumping index.
Green’s index
Green’s index (GI) is a modification of the index of cluster size that is independent of n the number of sample units.[64]
This index equals 0 if the distribution is random, 1 if it is maximally aggregated and -1 / ( nm - 1 ) if it is uniform.
The distribution of Green's index is not currently known so statistical tests have been difficult to devise for it.
If the population obeys Taylor's law
Binary dispersal index
Binary sampling (presence/absence) is frequently used where it is difficult to obtain accurate counts. The dispersal index (D) is used when the study population is divided into a series of equal samples ( number of units = N: number of units per sample = n: total population size = n x N ).[20] The theoretical variance of a sample from a population with a binomial distribution is
where s2 is the variance, n is the number of units sampled and p is the mean proportion of sampling units with at least one individual present. The dispersal index (D) is defined as the ratio of observed variance to the expected variance. In symbols
where varobs is the observed variance and varbin is the expected variance. The expected variance is calculated with the overall mean of the population. Values of D > 1 are considered to suggest aggregation. D( n - 1 ) is distributed as the chi squared variable with n - 1 degrees of freedom where n is the number of units sampled.
An alternative test is the C test.[65]
where D is the dispersal index, n is the number of units per sample and N is the number of samples. C is distributed normally. A statistically significant value of C indicates overdispersion of the population.
D is also related to intraclass correlation ( ρ ) which is defined as[66]
where T is the number of organisms per sample, p is the likelihood of the organism having the sought after property (diseased, pest free, etc), and xi is the number of organism in the ith unit with this property. T must be the same for all sampled units. In this case with n constant
If the data can be fitted with a beta-binomial distribution then[66]
where θ is the parameter of the distribution.[65]
Related statistics
A number of statistical tests are known that may be of use in applications.
de Oliveria's statistic
A related statistic suggested by de Oliveria[67] is the difference of the variance and the mean.[68] If the population is Poisson distributed then
where t is the Poisson parameter, s2 is the variance, m is the mean and n is the sample size. The expected value of s2 - m is zero. This statistic is distributed normally.[69]
If the Poisson parameter in this equation is estimated by putting t = m, after a little manipulation this statistic can be written
This is almost identical to Katz's statistic with ( n - 1 ) replacing n. Again OT is normally distributed with mean 0 and unit variance for large n.
- Note
de Oliveria actually suggested that the variance of s2 - m was ( 1 - 2t1/2 + 3t ) / n where t is the Poisson parameter. He suggested that t could be estimated by putting it equal to the mean (m) of the sample. Further investigation by Bohning[68] showed that this estimate of the variance was incorrect. Bohning's correction is given in the equations above.
Clapham's test
In 1936 Clapham proposed using the ratio of the variance to the mean as a test statistic (the relative variance).[70] In symbols
For a Possion distribution this ratio equals 1. To test for deviations from this value he prosed testing its value against the chi square distribution with n degrees of freedom where n is the number of sample units. The distribution of this statistic was studied further by Blackman[71] who noted that it was approximately normally distributed with a mean of 1 and a variance ( Vθ ) of
The derivation of the variance was re analysed by Bartlett[72] who considered it to be
For large samples these two formulae are in approximate agreement. This test is related to the later Katz's Jn statistic.
If the population obeys Taylor's law then
- Note
A refinement on this test has also been published[73] These authors noted that this test tends to detect overdispersion at higher scales even when this was not present in the data. They noted that that the use of the multinomial distribution may be more appropriate than the use of a Poisson distribution for such data. The statistic θ is distributed
where N is the number of sample units, n is the total number of samples examined and xi are the individual data values.
The expectation and variance of θ are
For large N E(θ) is approximately 1 and
If the number of individuals sampled ( n ) is large this estimate of the variance is in agreement with those derived earlier. However for smaller samples these latter estimates are more precise and should be used.
See also
- Morisita's overlap index
- Natural exponential family
- Scaling pattern of occupancy
- Spatial ecology
- Watson's power law
References
- ↑ Kilpatrick AM & Ives AR (2003) Species interactions can explain Taylor's power law for ecological time series. Nature 422, 65–68 doi:10.1038/nature01471
- ↑ Taylor LR (1961) Aggregation, variance and the mean. Nature 189, 732–735
- ↑ Ramsayer J, Fellous S, Cohen JE & Hochberg ME (2011) Taylor's Law holds in experimental bacterial populations but competition does not influence the slope. Biology Letters
- ↑ Anderson RM & May RM Nature (1989) 333, 514–519
- ↑ 5.0 5.1 Boag B, Hackett CA, Topham PB (1992) The use of Taylor's power law to describe the aggregated distribution of gastro-intestinal nematodes of sheep. Inter J Parasitol 22 (3) 267–270
- ↑ Kendal WS, Jørgensen B (2011) Taylor's power law and fluctuation scaling explained by a central-limit-like convergence. Phys Rev E Stat Nonlin Soft Matter Phys 83(6 Pt 2):066115
- ↑ Kendal WS (1995) A probabilistic model for the variance to mean power law in ecology. Ecological Modelling 80, 293-297
- ↑ Fronczak A, Fronczak P (2010) Origins of Taylor's power law for fluctuation scaling in complex systems. Phys Rev E Stat Nonlin Soft Matter Phys 81(6 Pt 2):066112.
- ↑ Smith H F (1938) An empirical law describing heterogeneity in the yield of agricultural crops. J Agric Sci 28: 1-23
- ↑ Fracker SB & Brischle HA (1944) Measuring the local distribution of Ribes. Ecology 25: 283-303
- ↑ Hayman BI & Lowe AD (1961) The transformation of counts of the cabbage aphid (Brevicovyne brassicae (L.)) NZ J Sci 4:271-278
- ↑ Southwood (1966) Ecological methods, with particular reference to the study of insect populations. Methuen (London)
- ↑ Rayner JMV (1985) Linear relations in biomechanics: the statistics of scaling functions. J Zoo 206 (3) 415–439
- ↑ Ferris H, Mullens TA, Foord KE (1990) Stability and characteristics of spatial description parameters for nematode populations. J Nematol 22(4):427-439
- ↑ Taylor LR, Taylor RAJ, Woiwod IP & Perry JN (1983) Behavioral dynamics. Nature 303:801–804
- ↑ Taylor LR & Woiwod IP (1980). Temporal stability as a density dependent species characteristic. J Anim Ecol 49: 209–224
- ↑ Wilson LT & Room PM (1982) The relative efficiency and reliability of three methods for sampling arthropods in Australian cotton fields. J Aust Entomol Soc 21: 175-181
- ↑ Banerjee B (1976) Variance to mean ratio and the spatial distribution of animals. Experientia 32: 993-994
- ↑ Hanski I (1982) On patterns of temporal and spatial variation in animal populations. Ann Zoo Fennici 19:21–37
- ↑ 20.0 20.1 Gottwald TR, Bassanezi RB, Amorim L, Bergamin-Filho A (2007) Spatial pattern analysis of citrus canker-infected plantings in São Paulo, Brazil, and augmentation of infection elicited by the Asian leafminer. Phytopathology 97(6):674-683
- ↑ Clark SJ & Perry JN (1994) Small sample estimation for Taylor's power law. Environment Ecol Stats 1 (4) 287–302, doi:10.1007/BF00469426
- ↑ 22.0 22.1 22.2 Wilson LT & Room PM (1983) Clumping patterns of fruit and arthropods in cotton with implications for binomial sampling. Environ Entomol 12:50-54
- ↑ 23.0 23.1 23.2 23.3 Bliss CI and Fisher RA (1953) Fitting the negative binomial distribution to biological data (also includes note on the efficient fitting of the negative binomial). Biometrics 9: 177-200
- ↑ Jones VP (1991) Binomial sampling plans for tentiform leafminer (Lepidoptera: Gracillariidae) on apple in Utah. J Econ Entomol 84: 484-488
- ↑ 25.0 25.1 Katz L (1965) United treatment of a broad class of discrete probability distributions. in Proceedings of the International Symposium on Discrete Distributions. Montreal
- ↑ Jewel W, Sundt B (1981) Improved approximations for the distribution of a heterogeneous risk portfolio. Bull Assoc Swiss Act 81: 221-240
- ↑ Foley P (1994) Predicting extinction times from environmental stochasticity and carrying capacity. Conserv Biol 8: 124-137
- ↑ Pertoldi C, Bach LA, Loeschcke V (2008) On the brink between extinction and persistence. Biol Direct 3: 47 doi: 10.1186/1745-6150-3-47
- ↑ Halley J, Inchausti P (2002) Lognormality in ecological time series. Oikos 99:518–530. doi: 10.1034/j.1600-0706.2002.11962.x
- ↑ Southwood TRE & Henderson PA (2000) Ecological methods. 3rd ed. Blackwood, Oxford
- ↑ Service MW (1971) Studies on sampling larval populations of the Anopheles gambiae complex. Bull World Health Organ 45: 169-180
- ↑ 32.0 32.1 32.2 Southwood TRE (1978) Ecological methods. Chapman & Hall, London, England
- ↑ Karandinos MG (1976) Optimum sample size and comments on some published formulae. Bull Entomol Soc Am 22:417-421
- ↑ Ruesink WG (1980) Introduction to sampling theory, in Kogan M & Herzog DC (eds.) Sampling Methods in Soybean Entomology. Springer-Verlag New York, Inc, New York. pp 61-78
- ↑ Bisseleua DHB, Yede, & Vida S (2011) Dispersion models and sampling of cacao mirid bug Sahlbergella singularis (Hemiptera: Miridae) on theobroma cacao in southern Cameroon. Environ Entomol 40(1): 111-119
- ↑ Wilson LT, Gonzalez D & Plant RE(1985) Predicting sampling frequency and economic status of spider mites on cotton. Proc. Beltwide Cotton Prod Res Conf, National Cotton Council of America, Memphis, TN pp 168-170
- ↑ Green RH (1970) On fixed precision level sequential sampling. Res Pop Ecol 12 (2) 249-251 doi:10.1007/BF02511568
- ↑ Myers JH (1978). Selecting a measure of dispersion. Environ Entomol 7: 619–621
- ↑ Bartlett MS (1936) J R Statist Soc, Supp 3: 185
- ↑ 40.0 40.1 Iwao S (1968) A new regression method for analyzing the aggregation pattern of animal populations. Res Popul Ecol 10: 1–20
- ↑ Nachman G (1981) A mathematical model of the functional relationship between density and spatial distribution of a population. J Anim Ecol 50: 453-460
- ↑ Kono T & Sugino T (1958) On the estimation of density of rice stem infested by the rice stem borer. Japanese J Appl Entomol and Zool 2: 184
- ↑ Binns MR & Bostonian NJ (1990) Robustness in empirically based binomial decision rules for integrated pest management. J Econ Entomol 83: 420-442
- ↑ 44.0 44.1 Nachman G (1984) Estimates of mean population density and spatial distribution of Tetranychus urticae (Acarina: Tetranychidae) and Phytoseiulus persimilis (Acarina: Phytoseiidae) based upon the proportion of empty sampling units. J Appl Ecolog 21: 903-991
- ↑ Schaalje GB ,Butts RA, Lysyk TL (1991) Simulation studies of binomial sampling: a new variance estimator and density pre&ctor, with special reference to the Russian wheat aphid (Homoptera: Aphididae). J Econ Entomol 84: 140-147
- ↑ Hughes G & Madden LV (1992) Aggregation and incidence of disease. Plant Pathol. 41:657-660.
- ↑ Madden LV & Hughes G (1995) Plant disease incidence: Distributions, heterogeneity, and temporal analysis. Annu. Rev. Phytopathol. 33:529-564
- ↑ Shiyomi M, Egawa T, Yamamoto Y (1998) Negative hypergeometric series and Taylor's power law in occurrence of plant populations in semi-natural grassland in Japan. Proceedings of the 18th International Grassland Congress on grassland management. The Inner Mongolia Univ Press pp35-43 (1998)
- ↑ Wilson L T & Room PM (1983) Clumping patterns of fruit and arthropods in cotton, with implications for binomial sampling. Environ Entomol 12: 50-54
- ↑ Perry JN & Taylor LR(1986). Stability of real interacting populations in space and time: implications, alternatives and negative binomial. J Animal Ecol 55: 1053–1068
- ↑ 51.0 51.1 Elliot JM (1977) Some methods for the statistical analysis of samples of benthic invertebrates. 2nd ed. Freshwater Biological Association, Cambridge, United Kingdom
- ↑ 52.0 52.1 52.2 52.3 Lloyd M (1967) Mean crowding. J Anim Ecol 36: 1-30
- ↑ Ifoulis AA, Savopoulou-Soultani M (2006) Developing optimum sample size and multistage sampling plans for Lobesia botrana (Lepidoptera: Tortricidae) larval infestation and injury in northern Greece. J Econ Entomol 99(5):1890–1898
- ↑ 54.0 54.1 Ho CC (1993) Dispersion statistics and sample size estimates for Tetranychus kanzawai (Acari: Tetranychidae) on mulberry. Environ Entomol 22: 21-25
- ↑ Iwao S (1975) A new method of sequential sampling to classify populations relative to a critical density. Res Popul Ecol 16: 281-28
- ↑ Kuno E (1969) A new method of sequential sampling to obtain the with a fixed level of precision. Res. Pooul. Ecol 11: 127-136
- ↑ Parrella MP, Jones VP (1985) Yellow traps as monitoring tools for Liriomyza trifolii (Diptera: Agromyzidae) in chrysanthemum greenhouses. J Econ Entomol 78, 53-56
- ↑ 58.0 58.1 Morisita M (1959) Measuring the dispersion and the analysis of distribution patterns. Memoires of the Faculty of Science, Kyushu University Series E. Biol 2:215-235
- ↑ Pedigo LP & Buntin GD (1994) Handbook of sampling methods for arthropods in agriculture. CRC Boca Raton FL
- ↑ Smith-Gill SJ (1975) Cytophysiological basis of disruptive pigmentary patterns in the leopard frog Rana pipiens. II. Wild type and mutant cell specific patterns. J Morphol 146, 35-54
- ↑ Fisher RA (1925) Statistical methods for research workers. Hafner, New York
- ↑ 62.0 62.1 Hoel P (1943) On the indices of dispersion. Ann Math Statist 14: 155
- ↑ David FN & Moore PG (1954) Notes on contagious distributions in plant populations. Ann Bot of London 18:47-53
- ↑ Green RH (1966) Measurement of non-randomness in spatial distributions. Res Pop Ecol 8:1-7
- ↑ 65.0 65.1 Hughes G & Madden LV (1993) Using the beta-binomial distribution to describe aggregated patterns of disease incidence. Phytopathology 83:759-763
- ↑ 66.0 66.1 Fleiss JL (1981) Statistical methods for rates and proportions. 2nd ed. Wiley, Newy York, USA
- ↑ de Oliveria T (1965) Some elementary tests for mixtures of discrete distributions, in Patil, GP ed., Classical and contagious discrete distributions. Calcutá, Calcutta Publishing Society pp379-384
- ↑ 68.0 68.1 Bohning D (1994) A note on a test for Poisson overdispersion. Biometrica 81(2) 418-419
- ↑ Ping S (1995) Further study on the statistical test to detect spatial pattern. Biometrical J 37(2) 199–203
- ↑ Clapham AR (1936) Overdispersion in grassland communities and the use of statistical methods in plant ecology. J Ecol 14: 232
- ↑ Blackman GE (1942) Statistical and ecological studies on the distribution of species in plant communities. I. Dispersion as a factor in the study of changes in plant populations. Ann Bot N.s. vi: 351
- ↑ Greig-Smith P (1952) The use of random and contiguous quadrats in the study of the structure of plant communities. Ann. Bot. 16:293-316
- ↑ Gosset E, Louis B (1986) The binning analysis - Towards a better significance test. Astrophysics Space Sci 120 (2) 263-306






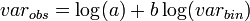
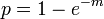



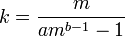

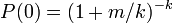

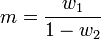

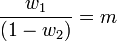
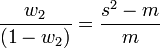
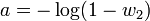

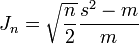



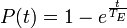
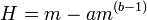
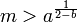
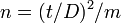
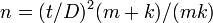
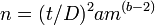

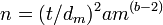






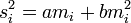




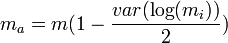



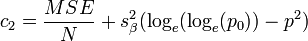
![c_{3}={\frac {\exp(a+(b-2)[\alpha -\beta \log _{e}(p_{0})])}{n}}](/2014-wikipedia_en_all_02_2014/I/media/c/c/a/f/ccafc294d9d31b1be50af3b49b9ee3a5.png)