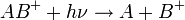Tandem mass spectrometry

Tandem mass spectrometry, also known as MS/MS or MS2, involves multiple steps of mass spectrometry selection, with some form of fragmentation occurring in between the stages.[2]
Tandem MS instruments
Multiple stages of mass analysis separation can be accomplished with individual mass spectrometer elements separated in space or using a single mass spectrometer with the MS steps separated in time.
Tandem in space
In tandem mass spectrometry in space, the separation elements are physically separated and distinct, although there is a physical connection between the elements to maintain high vacuum. These elements can be sectors, transmission quadrupole, or time-of-flight. When using multiple quadrupoles, they can act as both mass analyzers and collision chambers.
Tandem in time
By doing tandem mass spectrometry in time, the separation is accomplished with ions trapped in the same place, with multiple separation steps taking place over time. A quadrupole ion trap or FTMS instrument can be used for such an analysis. Trapping instruments can perform multiple steps of analysis, which is sometimes referred to as MSn (MS to the n). Often the number of steps, n, is not indicated, but occasionally the value is specified; for example MS3 indicates three stages of separation.
Notation
Schematic of tandem mass spectrometry
For tandem mass spectrometry in space, the different elements are often noted in shorthand.
- Q – Quadrupole mass analyzer
- q – Radio frequency collision quadrupole
- TOF – Time-of-flight mass analyzer
- B – Magnetic sector
- E – Electric sector
The notation can be combined to indicate various hybrid instrument, for example
- QqQ – Triple quadrupole mass spectrometer
- QTOF – Quadrupole time-of-flight mass spectrometer (also QqTOF)
- BEBE – Four-sector (reverse geometry) mass spectrometer
Tandem MS experiments
There are a number of different tandem MS experiments and all have their own applications and offer their own information. An instrument equipped for tandem MS can still be used to run MS experiments. Tandem MS can be done in either time or space. Tandem MS in space involves the physical separation of the instrument components (QqQ or QTOF), tandem MS in time involves the use of an ion trap.
Tandem MS modes
There are four main scan experiments possible using MS/MS:
- Precursor ion scan
- The product ion is selected in the second mass analyzer, and the precursor masses are scanned in the first mass analyzer. A precursor ion scan cannot be done with time based MS instruments. Note that precursor ion[3] is synonymous with parent ion[4] and product ion[5] with daughter ion;[6] however the use of these anthropomorphic terms is discouraged.[7][8]
- Product ion scan
- A precursor ion is selected in the first stage, allowed to fragment and then all resultant masses are scanned in the second mass analyzer and detected in the detector that is positioned after the second mass analyzer. This experiment is commonly performed to identify transitions used for quantification by tandem MS.
- Neutral loss scan
- The first mass analyzer scans all the masses. The second mass analyzer also scans, but at a set offset from the first mass analyzer.[9] This offset corresponds to a neutral loss that is commonly observed for the class of compounds. Neutral loss scans cannot be done with time based MS instruments. In a constant-neutral-loss scan, all precursors that undergo the loss of a specified common neutral are monitored. To obtain this information, both mass analyzers are scanned simultaneously, but with a mass offset that correlates with the mass of the specified neutral. Similar to the precursor-ion scan, this technique is also useful in the selective identification of closely related class of compounds in a mixture.
- Selected reaction monitoring
- Both mass analyzers are set to a selected mass. This mode is analogous to selected ion monitoring for MS experiments. A very selective analysis mode, which can increase sensitivity.[10]
Fragmentation in tandem mass spectrometry
Fragmentation of gas-phase ions is essential to tandem mass spectrometry and occurs between different stages of mass analysis. There are many methods used to fragment the ions and these can result in different types of fragmentation and thus different information about the structure and composition of the molecule.
In-source fragmentation
Often, the ionization process is sufficiently violent to leave the resulting ions with sufficient internal energy to fragment within the mass spectrometer. If the product ions persist in their non-equilibrium state for a moderate amount of time before auto-dissociation this process is called metastable fragmentation.[11][12] Nozzle-skimmer fragmentation refers to the purposeful induction of in-source fragmentation by increasing the nozzle-skimmer potential on usually electrospray based instruments. Although in-source fragmentation allows for fragmentation analysis, it is not technically tandem mass spectrometry unless metastable ions are mass analyzed or selected before auto-dissociation and a second stage of analysis is performed on the resulting fragments. In-source fragmentation is often used in addition to tandem mass spectrometry (with post-source fragmentation) to allow for two steps of fragmentation in a pseudo MS3-type of experiment.[13]
Post-source fragmentation
Post-source fragmentation is most often what is being used in a tandem mass spectrometry experiment. Energy can also be added to the ions, which are usually already vibrationally excited, through post-source collisions with neutral atoms or molecules, the absorption of radiation, or the transfer or capture of an electron by a multiply charged ion. Collision-induced dissociation (CID), also called collisionally activated dissociation (CAD), involves the collision of an ion with a neutral atom or molecule in the gas phase and subsequent dissociation of the ion.[14][15] For example, consider
where the ion 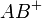 collides with the neutral species M and subsequently breaks apart. The details of this process are described by collision theory.
collides with the neutral species M and subsequently breaks apart. The details of this process are described by collision theory.
If an electron is added to a multiply charged positive ion, the Coulomb energy is liberated. Adding a free electron is called electron capture dissociation (ECD),[16] and is represented by
for a multiply protonated molecule M. Adding an electron through an ion-ion reaction is called electron transfer dissociation (ETD),[17] and is represented by
Such fragmentation can also occur with a deprotonated species, in which an electron is transferred from the specie to an cationic reagent in a negative electron transfer dissociation (NETD) event:
The energy required for dissociation can be added by photon absorption, resulting in ion photodissociation and represented by
where  represents the photon absorbed by the ion. Ultraviolet lasers can be used, but can lead to excessive fragmentation of biomolecules.[18] Infrared photons will heat the ions and cause dissociation if enough of them are absorbed. This process is called infrared multiphoton dissociation (IRMPD) and is often accomplished with a carbon dioxide laser and an ion trapping mass spectrometer such as a FTMS.[19] Blackbody radiation can also be used in a technique known as blackbody infrared radiative dissociation (BIRD).[20] In the BIRD method, the entire mass spectrometer vacuum chamber is heated to create infrared radiation.
represents the photon absorbed by the ion. Ultraviolet lasers can be used, but can lead to excessive fragmentation of biomolecules.[18] Infrared photons will heat the ions and cause dissociation if enough of them are absorbed. This process is called infrared multiphoton dissociation (IRMPD) and is often accomplished with a carbon dioxide laser and an ion trapping mass spectrometer such as a FTMS.[19] Blackbody radiation can also be used in a technique known as blackbody infrared radiative dissociation (BIRD).[20] In the BIRD method, the entire mass spectrometer vacuum chamber is heated to create infrared radiation.
With surface-induced dissociation (SID), the fragmentation is a result of the collision of an ion with a surface under high vacuum.[21][22]
Peptide fragmentation

A peptide sequence tag obtained by tandem mass spectrometry can be used to identify a peptide in a protein database.[23][24][25] A notation has been developed for indicating peptide fragments that arise from a tandem mass spectrum.[1] Peptide fragment ions are indicated by a, b, or c if the charge is retained on the N-terminus and by x, y or z if the charge is maintained on the C-terminus. The subscript indicates the number of amino acid residues in the fragment. Superscripts are sometimes used to indicate neutral losses in addition to the backbone fragmentation, * for loss of ammonia and ° for loss of water. Although peptide backbone cleavage is the most useful for sequencing and peptide identification other fragment ions may be observed under high energy dissociation conditions. These include the side chain loss ions d, v, w and immonium ions[26][27] and additional sequence-specific fragment ions associated with particular amino acid residues.[28][29]
Oligosaccharide fragmentation
Oligosaccharides may be sequenced using tandem mass spectrometry in a similar manner to peptide sequencing. Fragmentation generally occurs on either side of the glycosidic bond (b, c, y and z ions) but also under more energetic conditions through the sugar ring structure in a cross-ring cleavage (x ions). Again trailing subscripts are used to indicate position of the cleavage along the chain. For cross ring cleavage ions the nature of the cross ring cleavage is indicated by preceding superscripts.[30][31]
Oligonucleotide fragmentation
A notation for gas-phase fragmentation of oligonucleotide ions has been proposed.[32]
Newborn screening
Newborn screening is the process of testing newborn babies for treatable genetic, endocrinologic, metabolic and hematologic diseases.[33][34] The development of tandem mass spectrometry screening in the early 1990s led to a large expansion of potentially detectable congenital metabolic diseases that affect blood levels of organic acids.[35]
See also
- Accelerator mass spectrometry
- Bottom-up proteomics
- Charge remote fragmentation
- Cross section (physics)
- Hybrid mass spectrometer
- Mass-analyzed ion kinetic energy spectrometry
- Shotgun proteomics
- Top-down proteomics
- Triple quadrupole mass spectrometer
- Unimolecular ion decomposition
References
- ↑ 1.0 1.1 Roepstorff P, Fohlman J (1984). "Proposal for a common nomenclature for sequence ions in mass spectra of peptides". Biomed. Mass Spectrom. 11 (11): 601. doi:10.1002/bms.1200111109. PMID 6525415.
- ↑ IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "tandem mass spectrometer".
- ↑ IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "precursor ion".
- ↑ IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "parent ion".
- ↑ IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "product ion".
- ↑ IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "daughter ion".
- ↑ Bursey, Maurice M. (1991). "Comment to readers: Style and the lack of it". Mass Spectrometry Reviews 10: 1. doi:10.1002/mas.1280100102
- ↑ Adams, J. (1992). "To the editor". Journal of the American Society for Mass Spectrometry 3 (4): 473. doi:10.1016/1044-0305(92)87078-D
- ↑ Louris, John N.; Wright, Larry G.; Cooks, R. Graham.; Schoen, Alan E. (1985). "New scan modes accessed with a hybrid mass spectrometer". Analytical Chemistry 57 (14): 2918. doi:10.1021/ac00291a039
- ↑ deHoffman, Edmond; Stroobant, Vincent (2003). Mass Spectrometry: Principles and Applications. Toronto: Wiley. p. 133. ISBN 0-471-48566-7.
- ↑ IUPAC gold book definition of metastable ion (in mass spectrometry)
- ↑ IUPAC gold book definition of transient (chemical) species
- ↑ JAMS Vol. 7, Feb. 1996, pp 150-156
- ↑ Wells JM, McLuckey SA (2005). "Collision-induced dissociation (CID) of peptides and proteins". Meth. Enzymol. 402: 148–85. doi:10.1016/S0076-6879(05)02005-7. PMID 16401509.
- ↑ Sleno L, Volmer DA (2004). "Ion activation methods for tandem mass spectrometry". Journal of mass spectrometry : JMS 39 (10): 1091–112. doi:10.1002/jms.703. PMID 15481084.
- ↑ Cooper HJ, Håkansson K, Marshall AG (2005). "The role of electron capture dissociation in biomolecular analysis". Mass spectrometry reviews 24 (2): 201–22. doi:10.1002/mas.20014. PMID 15389856.
- ↑ Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF (2004). "Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry". Proc. Natl. Acad. Sci. U.S.A. 101 (26): 9528–33. Bibcode:2004PNAS..101.9528S. doi:10.1073/pnas.0402700101. PMC 470779. PMID 15210983.
- ↑ Morgan JW, Hettick JM, Russell DH (2005). "Peptide sequencing by MALDI 193-nm photodissociation TOF MS". Meth. Enzymol. 402: 186–209. doi:10.1016/S0076-6879(05)02006-9. PMID 16401510.
- ↑ Little DP, Speir JP, Senko MW, O'Connor PB, McLafferty FW (1994). "Infrared multiphoton dissociation of large multiply charged ions for biomolecule sequencing". Anal. Chem. 66 (18): 2809–15. doi:10.1021/ac00090a004. PMID 7526742.
- ↑ Schnier PD, Price WD, Jockusch RA, Williams ER (1996). "Blackbody Infrared Radiative Dissociation of Bradykinin and Its Analogues: Energetics, Dynamics, and Evidence for Salt-Bridge Structures in the Gas Phase". Journal of the American Chemical Society 118 (30): 7178–7189. doi:10.1021/ja9609157. PMC 1393282. PMID 16525512.
- ↑ Grill, Verena; Shen, Jianwei; Evans, Chris; Cooks, R. Graham (2001). "Collisions of ions with surfaces at chemically relevant energies: Instrumentation and phenomena". Review of Scientific Instruments 72 (8): 3149. Bibcode:2001RScI...72.3149G. doi:10.1063/1.1382641
- ↑ Mabud, M. (1985). "Surface-induced dissociation of molecular ions". International Journal of Mass Spectrometry and Ion Processes 67 (3): 285. doi:10.1016/0168-1176(85)83024-X
- ↑ Hardouin J (2007). "Protein sequence information by matrix-assisted laser desorption/ionization in-source decay mass spectrometry". Mass spectrometry reviews 26 (5): 672–82. doi:10.1002/mas.20142. PMID 17492750.
- ↑ Shadforth I, Crowther D, Bessant C (2005). "Protein and peptide identification algorithms using MS for use in high-throughput, automated pipelines". Proteomics 5 (16): 4082–95. doi:10.1002/pmic.200402091. PMID 16196103.
- ↑ Mørtz E, O'Connor PB, Roepstorff P, Kelleher NL, Wood TD, McLafferty FW, Mann M (1996). "Sequence tag identification of intact proteins by matching tanden mass spectral data against sequence data bases". Proc. Natl. Acad. Sci. U.S.A. 93 (16): 8264–7. Bibcode:1996PNAS...93.8264M. doi:10.1073/pnas.93.16.8264. PMC 38658. PMID 8710858.
- ↑ Richard S. Johnson, Stephen A. Martin and Klaus Biemann, Collision-induced fragmentation of (M + H)+ ions of peptides. Side chain specific sequence ions, International Journal of Mass Spectrometry and Ion Processes, Volume 86, 29 December 1988, Pages 137-154.
- ↑ A. M. Falick, W. M. Hines, K. F. Medzihradszky, M. A. Baldwin and B. W. Gibson, Low-mass ions produced from peptides by high-energy collision-induced dissociation in tandem mass spectrometry, Journal of the American Society for Mass Spectrometry, Volume 4, Issue 11, November 1993, Pages 882-893.
- ↑ Kevin M. Downard and Klaus Biemann, Amino acid sequence prerequisites for the formation of cn ion, Journal of the American Society for Mass Spectrometry Volume 4, Issue 11, November 1993, Pages 874–881.
- ↑ Kevin M. Downard and Klaus Biemann, Methionine specific sequence ions formed by the dissociation of protonated peptides at high collision energies, Journal of Mass Spectrometry, Volume 30, Issue 1, January 1995, pages 25–32.
- ↑ Bruno Domon, Catherine E Costello (1988). "A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates". Glycoconj. J. 5 (4): 397–409. doi:10.1007/BF01049915.
- ↑ Spina E, Cozzolino R, Ryan E, Garozzo D (2000). "Sequencing of oligosaccharides by collision-induced dissociation matrix-assisted laser desorption/ionization mass spectrometry". Journal of mass spectrometry : JMS 35 (8): 1042–8. doi:10.1002/1096-9888(200008)35:8<1042::AID-JMS33>3.0.CO;2-Y. PMID 10973004.
- ↑ J. Wu; S. A. McLuckey (2004). "Gas-phase fragmentation of oligonucleotide ions". International Journal of Mass Spectrometry 237 (2–3): 197. Bibcode:2004IJMSp.237..197W. doi:10.1016/j.ijms.2004.06.014
- ↑ Tarini BA (2007). "The current revolution in newborn screening: new technology, old controversies". Archives of pediatrics & adolescent medicine 161 (8): 767–72. doi:10.1001/archpedi.161.8.767. PMID 17679658.
- ↑ Kayton A (2007). "Newborn screening: a literature review". Neonatal network : NN 26 (2): 85–95. PMID 17402600.
- ↑ Chace DH, Kalas TA, Naylor EW (2003). "Use of tandem mass spectrometry for multianalyte screening of dried blood specimens from newborns". Clin. Chem. 49 (11): 1797–817. doi:10.1373/clinchem.2003.022178. PMID 14578311.
External links
Bibliography
- McLuckey, Scott A.; Busch, Kenneth L.; Glish, Gary L. (1988). Mass spectrometry/mass spectrometry: techniques and applications of tandem mass spectrometry. New York, N.Y: VCH Publishers. ISBN 0-89573-275-0.
- McLuckey, Scott A.; Glish, Gary L. Mass Spectrometry/Mass Spectrometry: Techniques and Applications of Tandem. Chichester: John Wiley & Sons. ISBN 0-471-18699-6.
- McLafferty, Fred W. (1983). Tandem mass spectrometry. New York: Wiley. ISBN 0-471-86597-4.
- Sherman, Nicholas E.; Kinter, Michael (2000). Protein sequencing and identification using tandem mass spectrometry. New York: John Wiley. ISBN 0-471-32249-0.
| ||||||||||||||||||||


![[M+nH]^{{n+}}+e^{-}\to {\bigg [}[M+(n-1)H]^{{(n-1)+}}{\bigg ]}^{*}\to fragments](/2014-wikipedia_en_all_02_2014/I/media/b/e/a/5/bea5d60c0444fbf8c6c739d50412db50.png)
![[M+nH]^{{n+}}+A^{-}\to {\bigg [}[M+(n-1)H]^{{(n-1)+}}{\bigg ]}^{*}+A\to fragments](/2014-wikipedia_en_all_02_2014/I/media/b/e/d/9/bed9e5f4a6b641eaec1b272b7427f65a.png)
![[M-nH]^{{n-}}+A^{+}\to {\bigg [}[M-nH]^{{(n+1)-}}{\bigg ]}^{*}+A\to fragments](/2014-wikipedia_en_all_02_2014/I/media/d/7/1/e/d71ee798d116037b343b974695029891.png)